orthodb
1、数据库

orthodb数据:
odb10v0_levels.tab.gz: NCBI taxonomy nodes where Ortho DB orthologous groups (OGs) are calculated
odb10v0_species.tab.gz: Ortho DB individual organism (aka species) ids based on NCBI taxonomy ids (mostly species level)
odb10v0_level2species.tab.gz: correspondence between level ids and species ids
odb10v0_genes.tab.gz: Ortho DB genes with some info
odb10v0_OGs.tab.gz: Ortho DB orthologous groups
odb10v0_OG2genes.tab.gz: OGs to genes correspondence
odb10v0_OG_xrefs.tab.gz: OG associations with GO, COG and InterPro ids
v9_v10_OGs_map.tab.gz mappings between the previous and current release orthologous group ids
odb10v0_fasta_<root>.tgz tar-ball with one fasta file per taxon id in the given root (bacteria,metazoa,fungi,plants)
2、odb10v0_levels.tab:
1. level NCBI taxonomy id
2. scientific name
3. total non-redundant count of genes in all underneath clustered species(在聚集的物种下面的所有的基因的总非重复计数)
4. total count of OGs built on it
5. total non-redundant count of species underneath

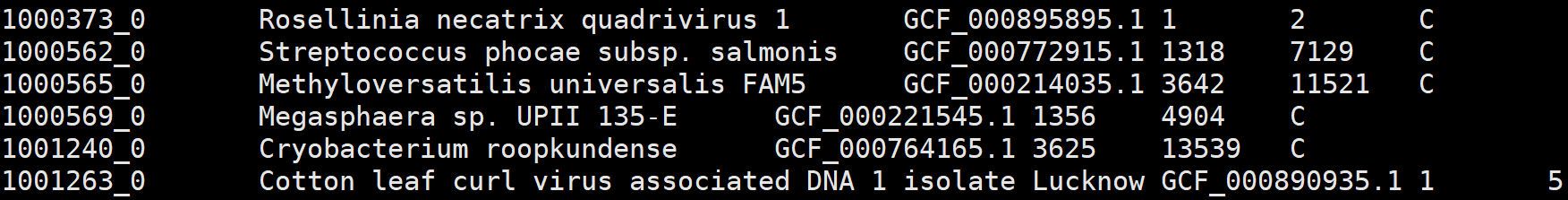
3、odb10v0_species.tab.gz
1. Ortho DB individual organism id, based on NCBI tax id
2. scientific name inherited from the most relevant NCBI tax id
3. genome asssembly id, when available
4. total count of clustered genes in this species
5. total count of the OGs it participates
6. mapping type, clustered(C) or mapped(M)
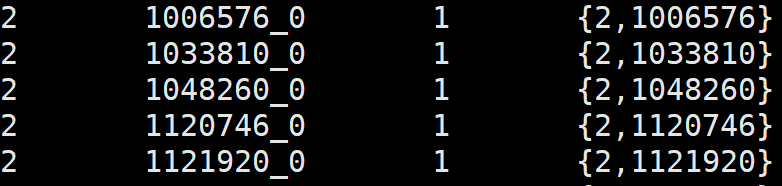
4、odb10v0_level2species.tab
1. top-most level NCBI tax id, one of [2,2157,2759,10239]
2. Ortho DB organism id
3. number of hops between the top-most level id and the NCBI tax id assiciated with the organism
4. ordered list of Ortho DB selected intermediate levels from the top-most level to the bottom one
5、odb10v0_genes.tab
1. Ortho DB unique gene id (not stable between releases)
2. organism tax id
3. protein original sequence id, as downloaded along with the sequence
4. Uniprot id, evaluated by mapping
5. ENSEMBL gene name, evaluated by mapping
6. NCBI gid, evaluated by mapping
7. description, evaluated by mapping
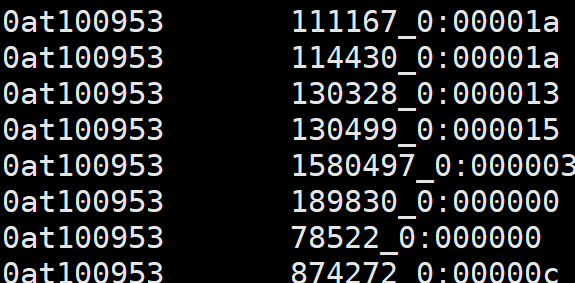
6、odb10v0_OG2genes.tab
1. OG unique id
2. Ortho DB gene id
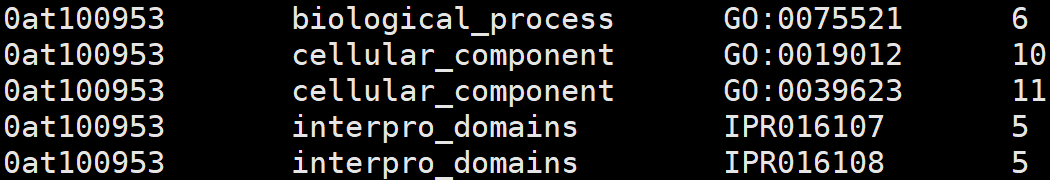
7、odb10v0_OG_xrefs.tab
1. OG unique id
2. external DB or DB section
3. external identifier
4. number of genes in the OG associated with the identifier
参考
https://www.orthodb.org/?page=filelist
orthodb的更多相关文章
- 【基因组预测】braker2基因结构注释要点记录
目录 流程使用 问题 记录下braker2的使用要点,以备忘记. 流程使用 braker2有很多流程,根据你的数据:组装的基因组.转录组.蛋白(同源,包括近缘或远缘)选择不同流程,官网有说明: htt ...
随机推荐
- Java的字符串
1.String 类 2.StringBuilder 类 1.String类 1.1.构造方法 String的构造方法格式 说明 new String(String st) 把字符串数据封装成字符串对 ...
- Warning: 执行完毕, 但带有警告 trigger trigger_EqPic_insert 已编译。
create or replace trigger trigger_EqPic_insert before insert on TB_EqPic for each row declare begin ...
- UVA-714-二分+贪心
题意:K个人复制M本书,每本书有Pi页,要求每个人都能分到至少一本书,如果分到多本书籍,分到的书籍编号是连续的,并且K个人里面分到的书籍总页数最大的那个人总数最小. 如果有多组解,保证 K1 < ...
- Jnlp
java web start解析jnlp文件,从网络宿主中,下载应用程序jar包,并运行. xxx.jnlp <?xml version="1.0" encoding=&qu ...
- Mysql 多字段去重
使用group by去重现在有如下表 id name age1 张三 232 李四 343 张三 234 李四 32 需求 : 按照name和age字段联合去重 sql如下 select * from ...
- openx -书表添加字段
OpenX的版本是2.8.10.在数据表加完数据库之后,还不能读取和保存字段. OpenX使用scheme来 管理数据库表和字段, 修改数据库结构同时也要修改相关schema, 一个是etc/tabl ...
- hadoop的块
http://hadoop.apache.org/docs/r1.0.4/cn/hdfs_design.html http://www.cnblogs.com/cloudma/articles/had ...
- Mybatis八( mybatis工作原理分析)
MyBatis的主要成员 Configuration MyBatis所有的配置信息都保存在Configuration对象之中,配置文件中的大部分配置都会存储到该类中 SqlSession ...
- pig简介
Apache Pig是MapReduce的一个抽象.它是一个工具/平台,用于分析较大的数据集,并将它们表示为数据流.Pig通常与 Hadoop 一起使用:我们可以使用Apache Pig在Hadoop ...
- kubectl version报did you specify the right host or port
现象: [root@localhost shell]# kubectl version Client Version: version.Info{Major:", GitVersion:&q ...
