POJ1080(LCS变形)
Human Gene Functions
Time Limit: 2000/1000 MS (Java/Others) Memory Limit: 65536/32768 K (Java/Others)
Total Submission(s): 2799 Accepted Submission(s): 1587
A human gene can be identified through a series of time-consuming biological experiments, often with the help of computer programs. Once a sequence of a gene is obtained, the next job is to determine its function. One of the methods for biologists to use in determining the function of a new gene sequence that they have just identified is to search a database with the new gene as a query. The database to be searched stores many gene sequences and their functions – many researchers have been submitting their genes and functions to the database and the database is freely accessible through the Internet.
A database search will return a list of gene sequences from the database that are similar to the query gene. Biologists assume that sequence similarity often implies functional similarity. So, the function of the new gene might be one of the functions that the genes from the list have. To exactly determine which one is the right one another series of biological experiments will be needed.
Your job is to make a program that compares two genes and determines their similarity as explained below. Your program may be used as a part of the database search if you can provide an efficient one.
Given two genes AGTGATG and GTTAG, how similar are they? One of the methods to measure the similarity of two genes is called alignment. In an alignment, spaces are inserted, if necessary, in appropriate positions of the genes to make them equally long and score the resulting genes according to a scoring matrix.
For example, one space is inserted into AGTGATG to result in AGTGAT-G, and three spaces are inserted into GTTAG to result in –GT--TAG. A space is denoted by a minus sign (-). The two genes are now of equal length. These two strings are aligned:
AGTGAT-G
-GT--TAG
In this alignment, there are four matches, namely, G in the second position, T in the third, T in the sixth, and G in the eighth. Each pair of aligned characters is assigned a score according to the following scoring matrix.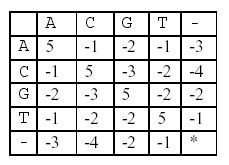
* denotes that a space-space match is not allowed. The score of the alignment above is (-3)+5+5+(-2)+(-3)+5+(-3)+5=9.
Of course, many other alignments are possible. One is shown below (a different number of spaces are inserted into different positions):
AGTGATG
-GTTA-G
This alignment gives a score of (-3)+5+5+(-2)+5+(-1) +5=14. So, this one is better than the previous one. As a matter of fact, this one is optimal since no other alignment can have a higher score. So, it is said that the similarity of the two genes is 14.
/*
ID: LinKArftc
PROG: 1080.cpp
LANG: C++
*/ #include <map>
#include <set>
#include <cmath>
#include <stack>
#include <queue>
#include <vector>
#include <cstdio>
#include <string>
#include <utility>
#include <cstdlib>
#include <cstring>
#include <iostream>
#include <algorithm>
using namespace std;
#define eps 1e-8
#define randin srand((unsigned int)time(NULL))
#define input freopen("input.txt","r",stdin)
#define debug(s) cout << "s = " << s << endl;
#define outstars cout << "*************" << endl;
const double PI = acos(-1.0);
const double e = exp(1.0);
const int inf = 0x3f3f3f3f;
const int INF = 0x7fffffff;
typedef long long ll; const int maxn = ;
int score[][] = {
{, -, -, -, -},
{-, , -, -, -},
{-, -, , -, -},
{-, -, -, , -},
{-, -, -, -, }
}; char ch_str1[maxn], ch_str2[maxn];
int str1[maxn], str2[maxn];
int dp[maxn][maxn]; int main() {
int T;
int len1, len2;
scanf("%d", &T);
while (T --) {
scanf("%d %s", &len1, ch_str1);
scanf("%d %s", &len2, ch_str2);
for (int i = ; i < len1; i ++) {
if (ch_str1[i] == 'A') str1[i] = ;
else if (ch_str1[i] == 'C') str1[i] = ;
else if (ch_str1[i] == 'G') str1[i] = ;
else if (ch_str1[i] == 'T') str1[i] = ;
else str1[i] = ;
}
for (int i = ; i < len2; i ++) {
if (ch_str2[i] == 'A') str2[i] = ;
else if (ch_str2[i] == 'C') str2[i] = ;
else if (ch_str2[i] == 'G') str2[i] = ;
else if (ch_str2[i] == 'T') str2[i] = ;
else str2[i] = ;
}
memset(dp, , sizeof(dp));
for (int i = ; i <= len1; i ++) dp[i][] = dp[i-][] + score[str1[i-]][];
for (int i = ; i <= len2; i ++) dp[][i] = dp[][i-] + score[][str2[i-]];
for (int i = ; i <= len1; i ++) {
for (int j = ; j <= len2; j ++) {
dp[i][j] = max(dp[i-][j-] + score[str1[i-]][str2[j-]],
max(dp[i][j-] + score[][str2[j-]], dp[i-][j] + score[str1[i-]][]));
}
}
printf("%d\n", dp[len1][len2]);
} return ;
}
POJ1080(LCS变形)的更多相关文章
- poj 1080 (LCS变形)
Human Gene Functions 题意: LCS: 设dp[i][j]为前i,j的最长公共序列长度: dp[i][j] = dp[i-1][j-1]+1;(a[i] == b[j]) dp[i ...
- POJ 1080( LCS变形)
题目链接: http://poj.org/problem?id=1080 Human Gene Functions Time Limit: 1000MS Memory Limit: 10000K ...
- UVA-1625-Color Length(DP LCS变形)
Color Length(UVA-1625)(DP LCS变形) 题目大意 输入两个长度分别为n,m(<5000)的颜色序列.要求按顺序合成同一个序列,即每次可以把一个序列开头的颜色放到新序列的 ...
- Advanced Fruits(HDU 1503 LCS变形)
Advanced Fruits Time Limit: 2000/1000 MS (Java/Others) Memory Limit: 65536/32768 K (Java/Others)T ...
- poj1080--Human Gene Functions(dp:LCS变形)
Human Gene Functions Time Limit: 1000MS Memory Limit: 10000K Total Submissions: 17206 Accepted: ...
- HDU 5791 Two ——(LCS变形)
感觉就是最长公共子序列的一个变形(虽然我也没做过LCS啦= =). 转移方程见代码吧.这里有一个要说的地方,如果a[i] == a[j]的时候,为什么不需要像不等于的时候那样减去一个dp[i-1][j ...
- uva 10723 Cyborg Genes(LCS变形)
题目:http://acm.hust.edu.cn/vjudge/contest/view.action?cid=107450#problem/C 题意:输入两个字符串,找一个最短的串,使得输入的两个 ...
- Combine String---hdu5727 &&& Zipper(LCS变形)
题目链接:http://poj.org/problem?id=2192 http://acm.split.hdu.edu.cn/showproblem.php?pid=5707 http://acm. ...
- HUST 4681 String (DP LCS变形)
题目链接:http://acm.hdu.edu.cn/showproblem.php?pid=4681 题目大意:给定三个字符串A,B,C 求最长的串D,要求(1)D是A的字序列 (2)D是B的子序列 ...
随机推荐
- Qt Qwdget 汽车仪表知识点拆解6 自定义控件
先贴上效果图,注意,没有写逻辑,都是乱动的 这里说一下控件自定义 图中标出的部分都是自定义的控件 这里如果我们有批量类似的功能,就可以使用自定义控件的方式,这里我已下面的自定义控件说一下,上面的在上一 ...
- 数据库sql命令
本文为转载,原文地址:http://www.cnblogs.com/cangqiongbingchen/p/4530333.html 1.说明:创建数据库CREATE DATABASE databas ...
- abs项目 - 战线拉的太长
abs项目 - 战线拉的太长 “从项目中来,到项目中去”. 坑是踩不完的,尽量做到不要踩重复的坑就好. 最近的这个项目,从2016的8月份左右开始立项,一直做到2017的2月份,还是有很多的问题在继续 ...
- nodeJs 调试异步程序追踪异步报错
DeprecationWarning: Calling an asynchronous function without callback is deprecated. 翻译: 不建议在不回调的情况下 ...
- DP入门(1)——数字三角形问题
一.问题描述 如上图所示,有一个由非负整数组成的三角形,第一行只有一个数,除了最下行之外每个数的左下方和右下方各有一个数.现请你在此数字三角形中寻找一条从首行到最下行的路径,使得路径上所经过的数字之和 ...
- 【WebService】——契约优先
相关博客: [WebService]--入门实例 [WebService]--SOAP.WSDL和UDDI 前言: 我们先来看一个契约优先的开发实例,通过熟悉他的开发流程,最后再和代码优先的方式进行比 ...
- [剑指Offer] 28.数组中出现次数超过一半的数字
[思路]将每个数字都存入map中作为key值,将它们出现的次数作为value值,当value超过一半时则返回其key值. class Solution { public: int MoreThanHa ...
- kali linux下的常用bash命令
虚拟机版本默认用户root 密码toor ls:显示当前目录包含的文件及文件夹 ls -l:以常规格式显示当前目录包含的文件及文件夹(开头字母解释:d:目录 -:文件 c:设备文件 l:链接 b:块设 ...
- BZOJ 3876:支线剧情(有下界最小费用最大流)
3876: [Ahoi2014]支线剧情 Description [故事背景]宅男JYY非常喜欢玩RPG游戏,比如仙剑,轩辕剑等等.不过JYY喜欢的并不是战斗场景,而是类似电视剧一般的充满恩怨情仇的剧 ...
- WebSocket简单介绍(WebSocket JavaScript 接口)(2)
上一节介绍了 WebSocket 规范,其中主要介绍了 WebSocket 的握手协议.握手协议通常是我们在构建 WebSocket 服务器端的实现和提供浏览器的WebSocket 支持时需要考虑的问 ...
