一元回归_R相关系数_多重检验
python机器学习-乳腺癌细胞挖掘(博主亲自录制视频)https://study.163.com/course/introduction.htm?courseId=1005269003&utm_campaign=commission&utm_source=cp-400000000398149&utm_medium=share
文件夹需要两个包


normality_check.py
# -*- coding: utf-8 -*-
'''
Author:Toby
QQ:231469242,all right reversed,no commercial use
normality_check.py
正态性检验脚本 ''' import scipy
from scipy.stats import f
import numpy as np
import matplotlib.pyplot as plt
import scipy.stats as stats
# additional packages
from statsmodels.stats.diagnostic import lillifors #正态分布测试
def check_normality(testData):
#20<样本数<50用normal test算法检验正态分布性
if 20<len(testData) <50:
p_value= stats.normaltest(testData)[1]
if p_value<0.05:
print"use normaltest"
print "data are not normal distributed"
return False
else:
print"use normaltest"
print "data are normal distributed"
return True #样本数小于50用Shapiro-Wilk算法检验正态分布性
if len(testData) <50:
p_value= stats.shapiro(testData)[1]
if p_value<0.05:
print "use shapiro:"
print "data are not normal distributed"
return False
else:
print "use shapiro:"
print "data are normal distributed"
return True if 300>=len(testData) >=50:
p_value= lillifors(testData)[1]
if p_value<0.05:
print "use lillifors:"
print "data are not normal distributed"
return False
else:
print "use lillifors:"
print "data are normal distributed"
return True if len(testData) >300:
p_value= stats.kstest(testData,'norm')[1]
if p_value<0.05:
print "use kstest:"
print "data are not normal distributed"
return False
else:
print "use kstest:"
print "data are normal distributed"
return True #对所有样本组进行正态性检验
def NormalTest(list_groups):
for group in list_groups:
#正态性检验
status=check_normality(group)
if status==False :
return False '''
group1=[2,3,7,2,6]
group2=[10,8,7,5,10]
group3=[10,13,14,13,15]
list_groups=[group1,group2,group3]
list_total=group1+group2+group3
#对所有样本组进行正态性检验
NormalTest(list_groups)
'''
correlalion_multiple.py
# -*- coding: utf-8 -*-
#斯皮尔曼等级相关(Spearman’s correlation coefficient for ranked data)
import math,pylab,scipy
import numpy as np
import scipy.stats as stats
from scipy.stats import t
from scipy.stats import f
import pandas as pd
import matplotlib.pyplot as plt
from statsmodels.stats.diagnostic import lillifors
import normality_check
import statsmodels.formula.api as sm
x=[40,42,50,55,65,78,84,100,116,125,130,140]
y=[130,150,155,140,150,154,165,170,167,180,175,185] list_group=[x,y]
sample=len(x)
#显著性
a=0.05 #数据可视化
plt.plot(x,y,'ro')
#斯皮尔曼等级相关,非参数检验
def Spearmanr(x,y):
print("use spearmanr,Nonparametric tests")
#样本不一致时,发出警告
if len(x)!=len(y):
print ("warming,the samples are not equal!")
r,p=stats.spearmanr(x,y)
print("spearman r**2:",r**2)
print("spearman p:",p)
if sample<500 and p>0.05:
print("when sample < 500,p has no mean(>0.05)")
print("when sample > 500,p has mean") #皮尔森 ,参数检验
def Pearsonr(x,y):
print("use Pearson,parametric tests")
r,p=stats.pearsonr(x,y)
print("pearson r**2:",r**2)
print("pearson p:",p)
if sample<30:
print("when sample <30,pearson has no mean") #皮尔森 ,参数检验,带有详细参数
def Pearsonr_details(x,y,xLabel,yLabel,formula):
n=len(x)
df=n-2
data=pd.DataFrame({yLabel:y,xLabel:x})
result = sm.ols(formula, data).fit()
print(result.summary()) #模型F分布显著性分析
print('\n')
print("linear relation Significant test:...................................")
#如果F检验的P值<0.05,拒绝H0,x和y无显著关系,H1成立,x和y有显著关系
if result.f_pvalue<0.05:
print ("P value of f test<0.05,the linear relation is right.") #R的显著检验
print('\n')
print("R significant test:...................................")
r_square=result.rsquared
r=math.sqrt(r_square)
t_score=r*math.sqrt(n-2)/(math.sqrt(1-r**2))
t_std=t.isf(a/2,df)
if t_score<-t_std or t_score>t_std:
print ("R is significant according to its sample size")
else:
print ("R is not significant") #残差分析
print('\n')
print("residual error analysis:...................................")
states=normality_check.check_normality(result.resid)
if states==True:
print("the residual error are normal distributed")
else:
print("the residual error are not normal distributed") #残差偏态和峰态
Skew = stats.skew(result.resid, bias=True)
Kurtosis = stats.kurtosis(result.resid, fisher=False,bias=True)
if round(Skew,1)==0:
print("residual errors normality Skew:in middle,perfect match")
elif round(Skew,1)>0:
print("residual errors normality Skew:close right")
elif round(Skew,1)<0:
print("residual errors normality Skew:close left") if round(Kurtosis,1)==3:
print("residual errors normality Kurtosis:in middle,perfect match")
elif round(Kurtosis,1)>3:
print("residual errors normality Kurtosis:more peak")
elif round(Kurtosis,1)<3:
print("residual errors normality Kurtosis:more flat") #自相关分析autocorrelation
print('\n')
print("autocorrelation test:...................................")
DW = np.sum( np.diff( result.resid.values )**2.0 )/ result.ssr
if round(DW,1)==2:
print("Durbin-Watson close to 2,there is no autocorrelation.OLS model works well")
else:
print("there may be autocorrelation")
#共线性检查
print('\n')
print("multicollinearity test:")
conditionNumber=result.condition_number
if conditionNumber>30:
print("conditionNumber>30,multicollinearity exists")
else:
print("conditionNumber<=30,multicollinearity not exists") #绘制残差图,用于方差齐性检验
Draw_residual(list(result.resid))
'''
result.rsquared
Out[28]: 0.61510660055413524
''' #kendalltau非参数检验
def Kendalltau(x,y):
print("use kendalltau,Nonparametric tests")
r,p=stats.kendalltau(x,y)
print("kendalltau r**2:",r**2)
print("kendalltau p:",p) #选择模型
def R_mode(x,y,xLabel,yLabel,formula):
#正态性检验
Normal_result=normality_check.NormalTest(list_group)
print ("normality result:",Normal_result)
if len(list_group)>2:
Kendalltau(x,y)
if Normal_result==False:
Spearmanr(x,y)
Kendalltau(x,y)
if Normal_result==True:
Pearsonr_details(x,y,xLabel,yLabel,formula) #调整的R方
def Adjust_Rsquare(r_square,n,k):
adjust_rSquare=1-((1-r_square)*(n-1)*1.0/(n-k-1))
return adjust_rSquare
'''
n=len(x)
n=10
k=1
r_square=0.615
Adjust_Rsquare(r_square,n,k)
Out[11]: 0.566875
''' #绘图
def Plot(x,y,yLabel,xLabel,Title):
plt.plot(x,y,'ro')
plt.ylabel(yLabel)
plt.xlabel(xLabel)
plt.title(Title)
plt.show() #绘图参数
yLabel='Alcohol'
xLabel='Tobacco'
Title='Sales in Several UK Regions'
Plot(x,y,yLabel,xLabel,Title)
formula='Alcohol ~ Tobacco' #绘制残点图
def Draw_residual(residual_list):
x=[i for i in range(1,len(residual_list)+1)]
y=residual_list
pylab.plot(x,y,'ro')
pylab.title("draw residual to check wrong number") # Pad margins so that markers don't get clipped by the axes,让点不与坐标轴重合
pylab.margins(0.3) #绘制网格
pylab.grid(True) pylab.show() R_mode(x,y,xLabel,yLabel,formula)
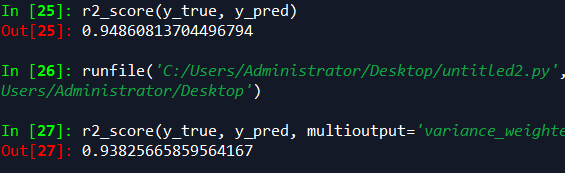
sklearn r平方计算
from sklearn.metrics import r2_score
y_true = [3, -0.5, 2, 7]
y_pred = [2.5, 0.0, 2, 8]
r2_score(y_true, y_pred) y_true = [[0.5, 1], [-1, 1], [7, -6]]
y_pred = [[0, 2], [-1, 2], [8, -5]]
r2_score(y_true, y_pred, multioutput='variance_weighted')
https://study.163.com/provider/400000000398149/index.htm?share=2&shareId=400000000398149( 欢迎关注博主主页,学习python视频资源,还有大量免费python经典文章)
一元回归_R相关系数_多重检验的更多相关文章
- Python_sklearn机器学习库学习笔记(一)_一元回归
一.引入相关库 %matplotlib inline import matplotlib.pyplot as plt from matplotlib.font_manager import FontP ...
- 一元回归1_基础(python代码实现)
python机器学习-乳腺癌细胞挖掘(博主亲自录制视频) https://study.163.com/course/introduction.htm?courseId=1005269003&u ...
- 机器学习(2):简单线性回归 | 一元回归 | 损失计算 | MSE
前文再续书接上一回,机器学习的主要目的,是根据特征进行预测.预测到的信息,叫标签. 从特征映射出标签的诸多算法中,有一个简单的算法,叫简单线性回归.本文介绍简单线性回归的概念. (1)什么是简单线性回 ...
- 标准方程法_岭回归_LASSO算法_弹性网
程序所用文件:https://files.cnblogs.com/files/henuliulei/%E5%9B%9E%E5%BD%92%E5%88%86%E7%B1%BB%E6%95%B0%E6%8 ...
- 零相关|回归|相关|相关系数|回归解释相关|r判断相关性|相关系数的区间估计|数据类型|非线性回归
零相关是什么? 零相关亦称“不相关”.相关的一种.两个变量的相关系数r=0时的相关.零相关表示两个变量非线性相关,这时两个变量可能相互独立,也可能曲线相关.对于正态变量,两个变量零相关与两个变量相互独 ...
- 一元回归_ols参数解读(推荐AAA)
sklearn实战-乳腺癌细胞数据挖掘(博客主亲自录制视频教程) https://study.163.com/course/introduction.htm?courseId=1005269003&a ...
- Linear regression with multiple variables(多特征的线型回归)算法实例_梯度下降解法(Gradient DesentMulti)以及正规方程解法(Normal Equation)
,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, ,, , ...
- R 分析回归(一元回归)
x <- c(,,,,,,,,,) # build X(predictor) y <- c(,,,,,,,,,) # build Y(dependent variable) mode(x) ...
- 回归分析法&一元线性回归操作和解释
用Excel做回归分析的详细步骤 一.什么是回归分析法 "回归分析"是解析"注目变量"和"因于变量"并明确两者关系的统计方法.此时,我们把因 ...
随机推荐
- Tensorflow、Pytorch、Keras的多GPU使用
Tensorflow.Pytorch.Keras的多GPU的并行操作 方法一 :使用深度学习工具提供的 API指定 1.1 Tesorflow tensroflow指定GPU的多卡并行的时候,也是可以 ...
- [数位DP]把枚举变成递推(未完)
动态规划(DP)是个很玄学的东西 数位DP实际上 就是把数字上的枚举变成按位的递推 有伪代码 for i =这一位起始值 i<=这一位终止值 dp[这一位][i]+=dp[这一位-1][i]+- ...
- 第一阶段android学习笔记
1.学习<第一行代码> 第一个android项目: 项目的注意点,如创建项目时包名具有唯一性,在做项目的时候要手动改成Project模式.还知道了引用字符串的两种方式. AS项目的三种依赖 ...
- 第15章 磁盘配额(Quota)与高级文件系统管理
磁盘配额(quota)的应用与实践 什么是quota 举例来说,用户的默认主文件夹是在/home下面,如果/home是个独立的分区,假设是10G,/home下有30个账号,这样30个用户共享这10G的 ...
- 【beta】阶段会议记录汇总
第一次: http://www.cnblogs.com/yumiaomiao/p/6026752.html 第二次: http://www.cnblogs.com/liquan/p/6031802.h ...
- PAT L1 - 056 猜数字
https://pintia.cn/problem-sets/994805046380707840/problems/994805074646122496 一群人坐在一起,每人猜一个 100 以内的数 ...
- HTTP协议 结构,get post 区别(阿里面试)
如果需要想了解相关的TCP的协议结构,底层架构,以及每次面试必问的三次握手,四次挥手可以 参考:TCP协议详解7层和4层解析(美团面试,阿里面试) 尤其是三次握手,四次挥手 具体发送的报文和状态都要掌 ...
- delphi 中如何执行SqlParameter形式的SQL语句
procedure TForm1.Button1Click(Sender: TObject); begin ADOConnection1.Open('); ADOQuery1.Close; ADOQu ...
- ZOJ3513_Human or Pig
这个题太坑爹了,题意也好纠结. 是这样的,给你一个n*m的矩形,中间有n*m个1*1的格子,有不同的跳跃方法.如果当前为human(人类)那么他可以有意识的选择自己下一步跳往何方:如果当前为pig(猪 ...
- 题解 P1059 【明明的随机数】
不会其他排序的小金羊又来水题了 本题我的思路:堆排,速度不需要算很快,AC就可以... 注意:初学者不宜抄此代码(压行严重) code: #include <cstdio> #includ ...