吴裕雄--天生自然 R语言数据可视化绘图(2)
par(ask=TRUE)
opar <- par(no.readonly=TRUE) # save original parameter settings library(vcd)
counts <- table(Arthritis$Improved)
counts
# Listing 6.1 - Simple bar plot
# vertical barplot
barplot(counts,
main="Simple Bar Plot",
xlab="Improvement", ylab="Frequency")
# horizontal bar plot
barplot(counts,
main="Horizontal Bar Plot",
xlab="Frequency", ylab="Improvement",
horiz=TRUE)
# obtain 2-way frequency table
library(vcd)
counts <- table(Arthritis$Improved, Arthritis$Treatment)
counts # Listing 6.2 - Stacked and grouped bar plots
# stacked barplot
barplot(counts,
main="Stacked Bar Plot",
xlab="Treatment", ylab="Frequency",
col=c("red", "yellow","green"),
legend=rownames(counts))
# grouped barplot
barplot(counts,
main="Grouped Bar Plot",
xlab="Treatment", ylab="Frequency",
col=c("red", "yellow", "green"),
legend=rownames(counts), beside=TRUE)
# Listing 6.3 - Bar plot for sorted mean values
states <- data.frame(state.region, state.x77)
means <- aggregate(states$Illiteracy, by=list(state.region), FUN=mean)
means means <- means[order(means$x),]
means barplot(means$x, names.arg=means$Group.1)
title("Mean Illiteracy Rate")

# Listing 6.3 - Bar plot for sorted mean values
states <- data.frame(state.region, state.x77)
means <- aggregate(states$Illiteracy, by=list(state.region), FUN=mean)
means means <- means[order(means$x),]
means barplot(means$x, names.arg=means$Group.1)
title("Mean Illiteracy Rate")

# Listing 6.4 - Fitting labels in bar plots
par(las=2) # set label text perpendicular to the axis
par(mar=c(5,8,4,2)) # increase the y-axis margin
counts <- table(Arthritis$Improved) # get the data for the bars # produce the graph
barplot(counts,
main="Treatment Outcome", horiz=TRUE, cex.names=0.8,
names.arg=c("No Improvement", "Some Improvement", "Marked Improvement")
)
par(opar)
# Spinograms
library(vcd)
attach(Arthritis)
counts <- table(Treatment,Improved)
spine(counts, main="Spinogram Example")
detach(Arthritis)

# Listing 6.5 - Pie charts
par(mfrow=c(2,2))
slices <- c(10, 12,4, 16, 8)
lbls <- c("US", "UK", "Australia", "Germany", "France") pie(slices, labels = lbls,
main="Simple Pie Chart")

pct <- round(slices/sum(slices)*100)
lbls <- paste(lbls, pct)
lbls <- paste(lbls,"%",sep="")
pie(slices,labels = lbls, col=rainbow(length(lbls)),
main="Pie Chart with Percentages")

library(plotrix)
pie3D(slices, labels=lbls,explode=0.1,
main="3D Pie Chart ") mytable <- table(state.region)
lbls <- paste(names(mytable), "\n", mytable, sep="")
pie(mytable, labels = lbls,
main="Pie Chart from a dataframe\n (with sample sizes)") par(opar)
mytable <- table(state.region)
lbls <- paste(names(mytable), "\n", mytable, sep="")
pie(mytable, labels = lbls,
main="Pie Chart from a dataframe\n (with sample sizes)") par(opar)

# Fan plots
library(plotrix)
slices <- c(10, 12,4, 16, 8)
lbls <- c("US", "UK", "Australia", "Germany", "France")
fan.plot(slices, labels = lbls, main="Fan Plot")
# Listing 6.6 - Histograms
# simple histogram 1
hist(mtcars$mpg)
# colored histogram with specified number of bins
hist(mtcars$mpg,
breaks=12,
col="red",
xlab="Miles Per Gallon",
main="Colored histogram with 12 bins")
# colored histogram with rug plot, frame, and specified number of bins
hist(mtcars$mpg,
freq=FALSE,
breaks=12,
col="red",
xlab="Miles Per Gallon",
main="Histogram, rug plot, density curve")
rug(jitter(mtcars$mpg))
lines(density(mtcars$mpg), col="blue", lwd=2)
# histogram with superimposed normal curve (Thanks to Peter Dalgaard)
x <- mtcars$mpg
h<-hist(x,
breaks=12,
col="red",
xlab="Miles Per Gallon",
main="Histogram with normal curve and box")
xfit<-seq(min(x),max(x),length=40)
yfit<-dnorm(xfit,mean=mean(x),sd=sd(x))
yfit <- yfit*diff(h$mids[1:2])*length(x)
lines(xfit, yfit, col="blue", lwd=2)
box()
# Listing 6.6 - Histograms
# simple histogram 1
hist(mtcars$mpg) # colored histogram with specified number of bins
hist(mtcars$mpg,
breaks=12,
col="red",
xlab="Miles Per Gallon",
main="Colored histogram with 12 bins") # colored histogram with rug plot, frame, and specified number of bins
hist(mtcars$mpg,
freq=FALSE,
breaks=12,
col="red",
xlab="Miles Per Gallon",
main="Histogram, rug plot, density curve")
rug(jitter(mtcars$mpg))
lines(density(mtcars$mpg), col="blue", lwd=2) # histogram with superimposed normal curve (Thanks to Peter Dalgaard)
x <- mtcars$mpg
h<-hist(x,
breaks=12,
col="red",
xlab="Miles Per Gallon",
main="Histogram with normal curve and box") xfit<-seq(min(x),max(x),length=40)
yfit<-dnorm(xfit,mean=mean(x),sd=sd(x))
yfit <- yfit*diff(h$mids[1:2])*length(x)
lines(xfit, yfit, col="blue", lwd=2)
box()
# Listing 6.7 - Kernel density plot
d <- density(mtcars$mpg) # returns the density data
plot(d) # plots the results

d <- density(mtcars$mpg)
plot(d, main="Kernel Density of Miles Per Gallon")
polygon(d, col="red", border="blue")
rug(mtcars$mpg, col="brown")
# Listing 6.8 - Comparing kernel density plots
par(lwd=2)
library(sm)
attach(mtcars) # create value labels
cyl.f <- factor(cyl, levels= c(4, 6, 8),
labels = c("4 cylinder", "6 cylinder", "8 cylinder")) # plot densities
sm.density.compare(mpg, cyl, xlab="Miles Per Gallon")
title(main="MPG Distribution by Car Cylinders")

# add legend via mouse click
colfill<-c(2:(2+length(levels(cyl.f))))
cat("Use mouse to place legend...","\n\n")
legend(locator(1), levels(cyl.f), fill=colfill)
detach(mtcars)
par(lwd=1)
# parallel box plots
boxplot(mpg~cyl,data=mtcars,
main="Car Milage Data",
xlab="Number of Cylinders",
ylab="Miles Per Gallon")
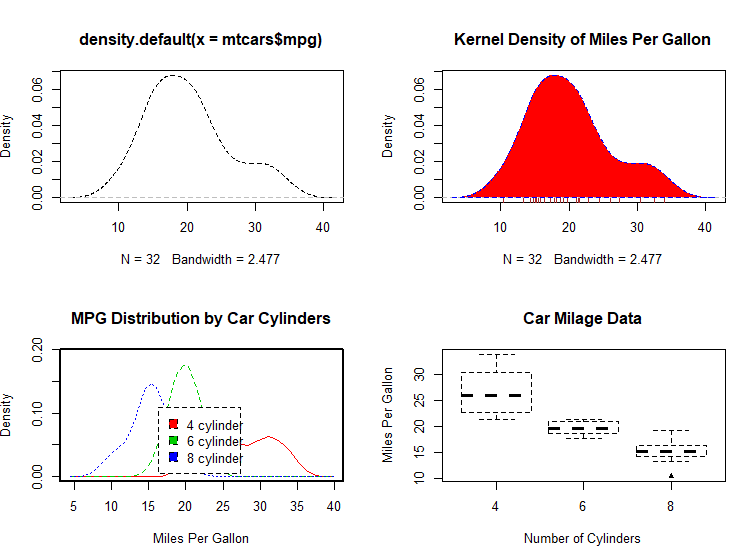
# notched box plots
boxplot(mpg~cyl,data=mtcars,
notch=TRUE,
varwidth=TRUE,
col="red",
main="Car Mileage Data",
xlab="Number of Cylinders",
ylab="Miles Per Gallon")

# Listing 6.9 - Box plots for two crossed factors
# create a factor for number of cylinders
mtcars$cyl.f <- factor(mtcars$cyl,
levels=c(4,6,8),
labels=c("4","6","8"))
# create a factor for transmission type
mtcars$am.f <- factor(mtcars$am,
levels=c(0,1),
labels=c("auto","standard"))
# generate boxplot
boxplot(mpg ~ am.f *cyl.f,
data=mtcars,
varwidth=TRUE,
col=c("gold", "darkgreen"),
main="MPG Distribution by Auto Type",
xlab="Auto Type")
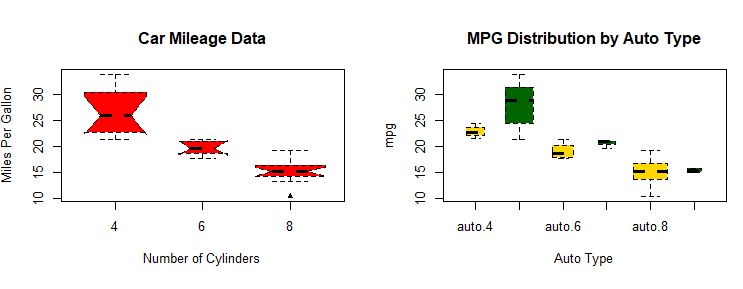
# Listing 6.10 - Violin plots library(vioplot)
x1 <- mtcars$mpg[mtcars$cyl==4]
x2 <- mtcars$mpg[mtcars$cyl==6]
x3 <- mtcars$mpg[mtcars$cyl==8]
vioplot(x1, x2, x3,
names=c("4 cyl", "6 cyl", "8 cyl"),
col="gold")
title("Violin Plots of Miles Per Gallon")
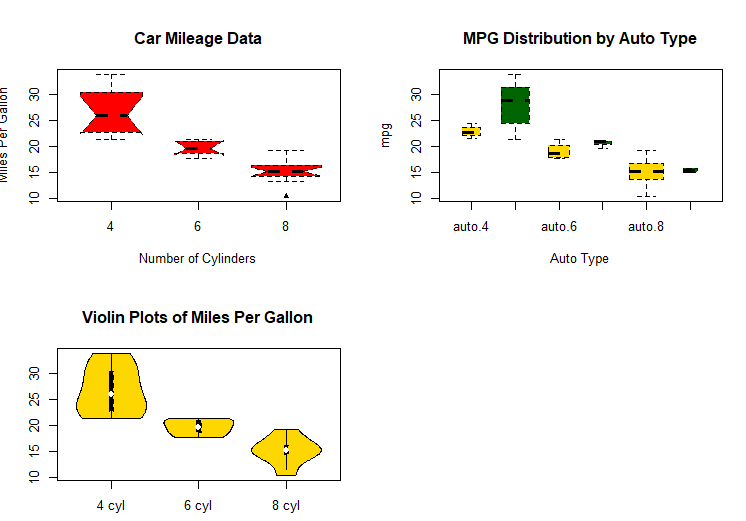
# dot chart
dotchart(mtcars$mpg,labels=row.names(mtcars),cex=.7,
main="Gas Mileage for Car Models",
xlab="Miles Per Gallon")
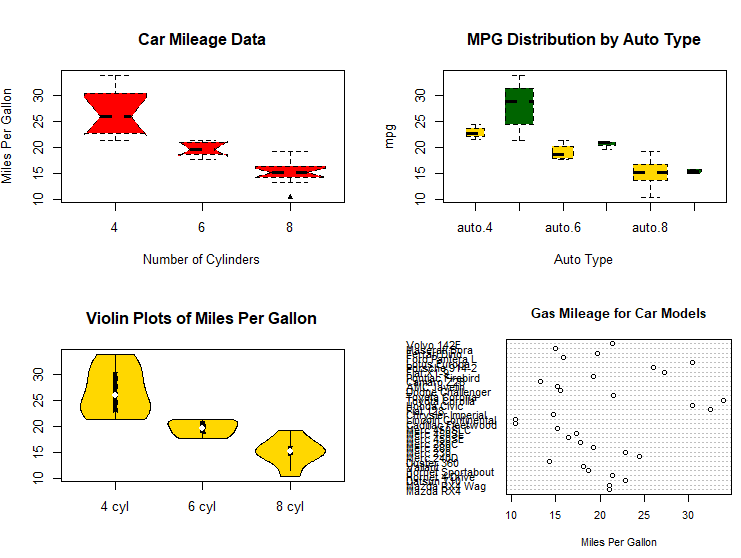
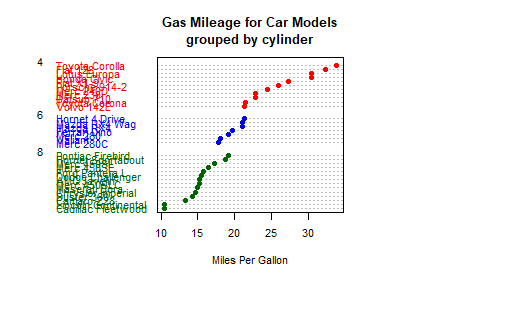
# Listing 6.11 - Dot plot grouped, sorted, and colored
x <- mtcars[order(mtcars$mpg),]
x$cyl <- factor(x$cyl)
x$color[x$cyl==4] <- "red"
x$color[x$cyl==6] <- "blue"
x$color[x$cyl==8] <- "darkgreen"
dotchart(x$mpg,
labels = row.names(x),
cex=.7,
pch=19,
groups = x$cyl,
gcolor = "black",
color = x$color,
main = "Gas Mileage for Car Models\ngrouped by cylinder",
xlab = "Miles Per Gallon")
吴裕雄--天生自然 R语言数据可视化绘图(2)的更多相关文章
- 吴裕雄--天生自然 R语言数据可视化绘图(3)
par(ask=TRUE) opar <- par(no.readonly=TRUE) # record current settings # Listing 11.1 - A scatter ...
- 吴裕雄--天生自然 R语言数据可视化绘图(4)
par(ask=TRUE) # Basic scatterplot library(ggplot2) ggplot(data=mtcars, aes(x=wt, y=mpg)) + geom_poin ...
- 吴裕雄--天生自然 R语言数据可视化绘图(1)
par(ask=TRUE) opar <- par(no.readonly=TRUE) # make a copy of current settings attach(mtcars) # be ...
- 吴裕雄--天生自然 R语言开发学习:R语言的安装与配置
下载R语言和开发工具RStudio安装包 先安装R
- 吴裕雄--天生自然 R语言开发学习:数据集和数据结构
数据集的概念 数据集通常是由数据构成的一个矩形数组,行表示观测,列表示变量.表2-1提供了一个假想的病例数据集. 不同的行业对于数据集的行和列叫法不同.统计学家称它们为观测(observation)和 ...
- 吴裕雄--天生自然 R语言开发学习:导入数据
2.3.6 导入 SPSS 数据 IBM SPSS数据集可以通过foreign包中的函数read.spss()导入到R中,也可以使用Hmisc 包中的spss.get()函数.函数spss.get() ...
- 吴裕雄--天生自然 R语言开发学习:处理缺失数据的高级方法(续一)
#-----------------------------------# # R in Action (2nd ed): Chapter 18 # # Advanced methods for mi ...
- 吴裕雄--天生自然 R语言开发学习:R语言的简单介绍和使用
假设我们正在研究生理发育问 题,并收集了10名婴儿在出生后一年内的月龄和体重数据(见表1-).我们感兴趣的是体重的分 布及体重和月龄的关系. 可以使用函数c()以向量的形式输入月龄和体重数据,此函 数 ...
- 吴裕雄--天生自然 R语言开发学习:使用键盘、带分隔符的文本文件输入数据
R可从键盘.文本文件.Microsoft Excel和Access.流行的统计软件.特殊格 式的文件.多种关系型数据库管理系统.专业数据库.网站和在线服务中导入数据. 使用键盘了.有两种常见的方式:用 ...
随机推荐
- Tornadofx学习笔记(2)——FxRecyclerView控件的打造
Tornadofx是基于javafx的一个kotlin框架,用来写些电脑版的小程序 基于Scroll Pane控件,仿造Android中的RecyclerView,实现的一款tornadofx的控件 ...
- Linux下安装Oracle后重启无法登录数据库ORA-01034:ORACLE not available
Linux下安装了数据库,安装完成后可以用,今天启动就不能用了,提示Oracle not available,后来查找资料,据说是oracle服务没有打开.如下方式可以解决问题. [root@root ...
- 面试总结 | Linux后台开发不得不看的知识点(给进军bat的你!)
目录 一 自我介绍 二 面试情况 三 相关知识点汇总 1 c/c++相关 2 计算机网络 3 数据结构相关 4 数据库相关 5 操作系统 6 Linux基础知识及应用编程(后台必备!) 7 大数问题 ...
- 物流跟踪API-快递单订阅
上一篇文章我们讲解了轨迹查询的接口,通过快递鸟接口可以实现实时查询物流轨迹,这次给大家推荐订阅服务功能. 为了更好的理解订阅服务,我们来做个对比, 即时查询是主动查询物流轨迹,需要我们主动调用接口才能 ...
- 时序数据库 Apache-IoTDB 源码解析之系统架构(二)
上一章聊到时序数据是什么样,物联网行业中的时序数据的特点:存量数据大.新增数据多(采集频率高.设备量多).详情请见: 时序数据库 Apache-IoTDB 源码解析之前言(一) 打一波广告,欢迎大家访 ...
- JavaScript 与 Java 有什么不同?
JavaScript 编程语言是由 Netscape,Inc. 开发的,它并不是 Java 平台的一部分. JavaScript 不会创建小应用程序或独立应用程序.在最常见的形式中,JavaScrip ...
- python笔记带你走向测试开发之路-第一篇(数据类型之数字,序列)
数字 数字的类型 数字是 Python中比较常用的数据类型,数字有可以分为: 整型 int如 1,2,3 浮点型 float如 2.1,3.5 长整型 long如 3L,需要注意的是 Python2. ...
- EMC networker nmm can restore and recover sqlserver as different name to different location
EMC networker nmm can restore and recover sqlserver as different name to different location That is ...
- android 基础学习笔记2
1.容器布局 一.线性布局 (LineaLayout) 方向:orientation =vertical / horizontal 重力(对齐) :gravity =bottom/right/left ...
- 【机器学习】算法原理详细推导与实现(六):k-means算法
[机器学习]算法原理详细推导与实现(六):k-means算法 之前几个章节都是介绍有监督学习,这个章解介绍无监督学习,这是一个被称为k-means的聚类算法,也叫做k均值聚类算法. 聚类算法 在讲监督 ...
