[Scikit-learn] 2.1 Clustering - Gaussian mixture models & EM
原理请观良心视频:机器学习课程 Expectation Maximisation

Expectation-maximization is a well-founded statistical algorithm to get around this problem by an iterative process.
- First one assumes random components (randomly centered on data points, learned from k-means, or even just normally distributed around the origin) and computes for each point a probability of being generated by each component of the model.
- Then, one tweaks the parameters to maximize the likelihood of the data given those assignments. Repeating this process is guaranteed to always converge to a local optimum.
实战:
X_train
Out[79]:
array([[ 4.3, 3. , 1.1, 0.1],
[ 5.8, 4. , 1.2, 0.2],
[ 5.7, 4.4, 1.5, 0.4],
...,
[ 6.5, 3. , 5.2, 2. ],
[ 6.2, 3.4, 5.4, 2.3],
[ 5.9, 3. , 5.1, 1.8]]) X_train.size
Out[80]: 444 classifier.means_
Out[81]:
array([[ 5.04594595, 3.45135126, 1.46486501, 0.25675684], # 1st 4d Gaussian
[ 5.92023012, 2.75827264, 4.42168189, 1.43882194], # 2nd 4d Gaussian
[ 6.8519452 , 3.09214071, 5.71373857, 2.0934678 ]]) # 3rd 4d Gaussian
classifier.covars_
Out[82]:
array([[ 0.08532076, 0.08532076, 0.08532076, 0.08532076],
[ 0.14443088, 0.14443088, 0.14443088, 0.14443088],
[ 0.1758563 , 0.1758563 , 0.1758563 , 0.1758563 ]])
本有四个变量,如何画在平面图上的呢?以上只取了前两维数据做图。
"""
==================
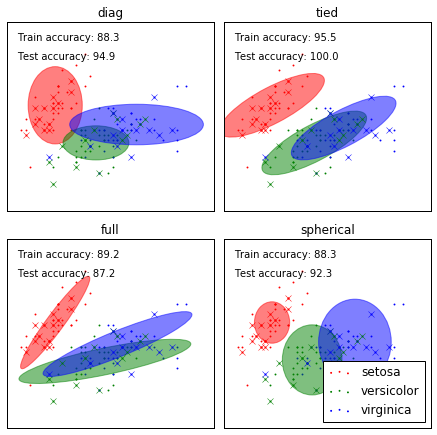
GMM classification
================== Demonstration of Gaussian mixture models for classification. See :ref:`gmm` for more information on the estimator. Plots predicted labels on both training and held out test data using a
variety of GMM classifiers on the iris dataset. Compares GMMs with spherical, diagonal, full, and tied covariance
matrices in increasing order of performance. Although one would
expect full covariance to perform best in general, it is prone to
overfitting on small datasets and does not generalize well to held out
test data. On the plots, train data is shown as dots, while test data is shown as
crosses. The iris dataset is four-dimensional. Only the first two
dimensions are shown here, and thus some points are separated in other
dimensions.
"""
print(__doc__) # Author: Ron Weiss <ronweiss@gmail.com>, Gael Varoquaux
# License: BSD 3 clause # $Id$ import matplotlib.pyplot as plt
import matplotlib as mpl
import numpy as np from sklearn import datasets
from sklearn.cross_validation import StratifiedKFold
from sklearn.externals.six.moves import xrange
from sklearn.mixture import GMM def make_ellipses(gmm, ax):
for n, color in enumerate('rgb'):
v, w = np.linalg.eigh(gmm._get_covars()[n][:2, :2])
u = w[0] / np.linalg.norm(w[0])
angle = np.arctan2(u[1], u[0])
angle = 180 * angle / np.pi # convert to degrees
v *= 9
ell = mpl.patches.Ellipse(gmm.means_[n, :2], v[0], v[1], 180 + angle, color=color)
ell.set_clip_box(ax.bbox)
ell.set_alpha(0.5)
ax.add_artist(ell)
iris = datasets.load_iris()
#数据预处理
# Break up the dataset into non-overlapping training (75%) and testing
# (25%) sets.
# 分层交叉验证,使得交叉验证抽到的样本符合原始样本的比例。
skf = StratifiedKFold(iris.target, n_folds=4)
# Only take the first fold.
train_index, test_index = next(iter(skf))
# next(iter())逐个遍历skf的elem, len(skf) = 4
# 随机获取了四组中的一组数据
X_train = iris.data [train_index]
y_train = iris.target[train_index]
X_test = iris.data [test_index]
y_test = iris.target[test_index]
#GMM初始化
n_classes = len(np.unique(y_train))
# y_train就三种值,代表有仨个Gaussian # Try GMMs using different types of covariances.
# 四种不同的type做GMM,然后存放在dict中
classifiers = dict((covar_type,
GMM(n_components=n_classes, covariance_type=covar_type, init_params='wc', n_iter=20)
)
for covar_type in ['spherical', 'diag', 'tied', 'full']
)
# NB:covar_type的表现往往体现在高斯分布图像的旋转
n_classifiers = len(classifiers) plt.figure(figsize=(3 * n_classifiers / 2, 6))
plt.subplots_adjust(bottom=.01, top=0.95, hspace=.15, wspace=.05,
left=.01, right=.99) for index, (name, classifier) in enumerate(classifiers.items()):
"""
dict_items([('diag', GMM(covariance_type='diag', init_params='wc', min_covar=0.001, n_components=5, n_init=1, n_iter=20, params='wmc', random_state=None, tol=0.001, verbose=0)),
('tied', GMM(covariance_type='tied', init_params='wc', min_covar=0.001, n_components=5, n_init=1, n_iter=20, params='wmc', random_state=None, tol=0.001, verbose=0)),
('full', GMM(covariance_type='full', init_params='wc', min_covar=0.001, n_components=5, n_init=1, n_iter=20, params='wmc', random_state=None, tol=0.001, verbose=0)),
('spherical', GMM(covariance_type='spherical', init_params='wc', min_covar=0.001, n_components=5, n_init=1, n_iter=20, params='wmc', random_state=None, tol=0.001, verbose=0))])
"""
# 数据训练
# Since we have class labels for the training data, we can
# initialize the GMM parameters in a supervised manner.
classifier.means_ = np.array([X_train[y_train == i].mean(axis=0) for i in xrange(n_classes)])
# axis=0 沿着Matrix的‘行’求统计量,NB:每个向量的第一元素求mean,第二个元素求mean ...
# Train the other parameters using the EM algorithm.
classifier.fit(X_train)
# 数据表现
h = plt.subplot(2, n_classifiers / 2, index + 1)
make_ellipses(classifier, h) for n, color in enumerate('rgb'):
data = iris.data[iris.target == n]
plt.scatter(data[:, 0], data[:, 1], 0.8, color=color,
label=iris.target_names[n])
# Plot the test data with crosses
for n, color in enumerate('rgb'):
data = X_test[y_test == n]
plt.plot(data[:, 0], data[:, 1], 'x', color=color) y_train_pred = classifier.predict(X_train)
train_accuracy = np.mean(y_train_pred.ravel() == y_train.ravel()) * 100
plt.text(0.05, 0.9, 'Train accuracy: %.1f' % train_accuracy,
transform=h.transAxes)
test_accuracy = np.mean(y_test_pred.ravel() == y_test.ravel()) * 100
plt.text(0.05, 0.8, 'Test accuracy: %.1f' % test_accuracy,
transform=h.transAxes) plt.xticks(())
plt.yticks(())
plt.title(name) plt.legend(loc='lower right', prop=dict(size=12)) plt.show()
New api: mixture.GMM
"""
=============================================
Density Estimation for a mixture of Gaussians
============================================= Plot the density estimation of a mixture of two Gaussians. Data is
generated from two Gaussians with different centers and covariance
matrices.
""" import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
from sklearn import mixture n_samples = 300 # generate random sample, two components
np.random.seed(0) # generate spherical data centered on (20, 20)
shifted_gaussian = np.random.randn(n_samples, 2) + np.array([20, 20]) # generate zero centered stretched Gaussian data
C = np.array([[0., -0.7], [3.5, .7]])
stretched_gaussian = np.dot(np.random.randn(n_samples, 2), C) # concatenate the two datasets into the final training set
X_train = np.vstack([shifted_gaussian, stretched_gaussian]) # fit a Gaussian Mixture Model with two components
clf = mixture.GMM(n_components=2, covariance_type='full')
clf.fit(X_train) # display predicted scores by the model as a contour plot
x = np.linspace(-20.0, 30.0)
y = np.linspace(-20.0, 40.0)
X, Y = np.meshgrid(x, y)
XX = np.array([X.ravel(), Y.ravel()]).T
Z = -clf.score_samples(XX)[0]
Z = Z.reshape(X.shape) CS = plt.contour(X, Y, Z, norm=LogNorm(vmin=1.0, vmax=1000.0),
levels=np.logspace(0, 3, 10))
CB = plt.colorbar(CS, shrink=0.8, extend='both')
plt.scatter(X_train[:, 0], X_train[:, 1], .8) plt.title('Negative log-likelihood predicted by a GMM')
plt.axis('tight')
plt.show()
如何判定模型中有几个Gaussian,Selecting the number of components in a classical GMM
The BIC criterion can be used to select the number of components in a GMM in an efficient way. In theory, it recovers the true number of components only in the asymptotic regime (i.e. if much data is available).
Note that using a DPGMM avoids the specification of the number of components for a Gaussian mixture model.
(NOTE:DPGMM会放在Dirichlet Process章节中学习)
哪个模型更加的好呢?目前常用有如下方法:
AIC = -2 ln(L) + 2k Akaike information criterion
BIC = -2 ln(L) + ln(n)*k Bayesian information criterion
HQ = -2 ln(L) + ln(ln(n))*k Hannan-quinn criterion
其中L是在该模型下的最大似然,n是数据数量,k是模型的变量个数。
注意这些规则只是刻画了用某个模型之后相对“真实模型”的信息损失【因为不知道真正的模型是什么样子,所以训练得到的所有模型都只是真实模型的一个近似模型】,所以用这些规则不能说明某个模型的精确度,即三个模型A, B, C,在通过这些规则计算后,我们知道B模型是三个模型中最好的,但是不能保证B这个模型就能够很好地刻画数据,因为很有可能这三个模型都是非常糟糕的,B只是烂苹果中的相对好的苹果而已。
这些规则理论上是比较漂亮的,但是实际在模型选择中应用起来还是有些困难的,例如上面我们说了5个变量就有32个变量组合,如果是10个变量呢?2的10次方,我们不可能对所有这些模型进行一一验证AIC, BIC,HQ规则来选择模型,工作量太大。
"""
=================================
Gaussian Mixture Model Selection
================================= This example shows that model selection can be performed with
Gaussian Mixture Models using information-theoretic criteria (BIC).
Model selection concerns both the covariance type
and the number of components in the model.
In that case, AIC also provides the right result (not shown to save time),
but BIC is better suited if the problem is to identify the right model.
Unlike Bayesian procedures, such inferences are prior-free. In that case, the model with 2 components and full covariance
(which corresponds to the true generative model) is selected.
"""
print(__doc__) import itertools import numpy as np
from scipy import linalg
import matplotlib.pyplot as plt
import matplotlib as mpl from sklearn import mixture # Number of samples per component
n_samples = 500 # Generate random sample, two components
np.random.seed(0)
C = np.array([[0., -0.1], [1.7, .4]])
X = np.r_[np.dot(np.random.randn(n_samples, 2), C),
.7 * np.random.randn(n_samples, 2) + np.array([-6, 3])] lowest_bic = np.infty
bic = []
n_components_range = range(1, 7)
cv_types = ['spherical', 'tied', 'diag', 'full']
for cv_type in cv_types:
for n_components in n_components_range:
# Fit a mixture of Gaussians with EM
gmm = mixture.GMM(n_components=n_components, covariance_type=cv_type)
gmm.fit(X)
bic.append(gmm.bic(X))
if bic[-1] < lowest_bic:
lowest_bic = bic[-1]
best_gmm = gmm
# 这里不需要 test set
bic = np.array(bic)
color_iter = itertools.cycle(['k', 'r', 'g', 'b', 'c', 'm', 'y'])
clf = best_gmm
bars = [] # Plot the BIC scores
spl = plt.subplot(2, 1, 1)
for i, (cv_type, color) in enumerate(zip(cv_types, color_iter)):
xpos = np.array(n_components_range) + .2 * (i - 2)
bars.append(plt.bar(xpos, bic[i * len(n_components_range):
(i + 1) * len(n_components_range)],
width=.2, color=color))
plt.xticks(n_components_range)
plt.ylim([bic.min() * 1.01 - .01 * bic.max(), bic.max()])
plt.title('BIC score per model')
xpos = np.mod(bic.argmin(), len(n_components_range)) + .65 +\
.2 * np.floor(bic.argmin() / len(n_components_range))
plt.text(xpos, bic.min() * 0.97 + .03 * bic.max(), '*', fontsize=14)
spl.set_xlabel('Number of components')
spl.legend([b[0] for b in bars], cv_types) # Plot the winner
splot = plt.subplot(2, 1, 2)
Y_ = clf.predict(X)
for i, (mean, covar, color) in enumerate(zip(clf.means_, clf.covars_,
color_iter)):
v, w = linalg.eigh(covar)
if not np.any(Y_ == i):
continue
plt.scatter(X[Y_ == i, 0], X[Y_ == i, 1], .8, color=color) # Plot an ellipse to show the Gaussian component
angle = np.arctan2(w[0][1], w[0][0])
angle = 180 * angle / np.pi # convert to degrees
v *= 4
ell = mpl.patches.Ellipse(mean, v[0], v[1], 180 + angle, color=color)
ell.set_clip_box(splot.bbox)
ell.set_alpha(.5)
splot.add_artist(ell) plt.xlim(-10, 10)
plt.ylim(-3, 6)
plt.xticks(())
plt.yticks(())
plt.title('Selected GMM: full model, 2 components')
plt.subplots_adjust(hspace=.35, bottom=.02)
plt.show()
[Scikit-learn] 2.1 Clustering - Gaussian mixture models & EM的更多相关文章
- [OpenCV] Samples 15: Background Subtraction and Gaussian mixture models
不错的草稿.但进一步处理是必然的,也是难点所在. Extended: 固定摄像头,采用Gaussian mixture models对背景建模. OpenCV 中实现了两个版本的高斯混合背景/前景分割 ...
- Gaussian Mixture Models and the EM algorithm汇总
Gaussian Mixture Models and the EM algorithm汇总 作者:凯鲁嘎吉 - 博客园 http://www.cnblogs.com/kailugaji/ 1. 漫谈 ...
- [Scikit-learn] 2.1 Clustering - Variational Bayesian Gaussian Mixture
最重要的一点是:Bayesian GMM为什么拟合的更好? PRML 这段文字做了解释: Ref: http://freemind.pluskid.org/machine-learning/decid ...
- 漫谈 Clustering (3): Gaussian Mixture Model
上一次我们谈到了用 k-means 进行聚类的方法,这次我们来说一下另一个很流行的算法:Gaussian Mixture Model (GMM).事实上,GMM 和 k-means 很像,不过 GMM ...
- 基于图嵌入的高斯混合变分自编码器的深度聚类(Deep Clustering by Gaussian Mixture Variational Autoencoders with Graph Embedding, DGG)
基于图嵌入的高斯混合变分自编码器的深度聚类 Deep Clustering by Gaussian Mixture Variational Autoencoders with Graph Embedd ...
- [zz] 混合高斯模型 Gaussian Mixture Model
聚类(1)——混合高斯模型 Gaussian Mixture Model http://blog.csdn.net/jwh_bupt/article/details/7663885 聚类系列: 聚类( ...
- Fisher Vector Encoding and Gaussian Mixture Model
一.背景知识 1. Discriminant Learning Algorithms(判别式方法) and Generative Learning Algorithms(生成式方法) 现在常见的模式 ...
- (原创)(三)机器学习笔记之Scikit Learn的线性回归模型初探
一.Scikit Learn中使用estimator三部曲 1. 构造estimator 2. 训练模型:fit 3. 利用模型进行预测:predict 二.模型评价 模型训练好后,度量模型拟合效果的 ...
- 聚类之高斯混合模型(Gaussian Mixture Model)【转】
k-means应该是原来级别的聚类方法了,这整理下一个使用后验概率准确评测其精度的方法—高斯混合模型. 我们谈到了用 k-means 进行聚类的方法,这次我们来说一下另一个很流行的算法:Gaussia ...
随机推荐
- bluetooth在linux应用开发
linux内Bluetooth的协议栈为BlueZ,http://www.bluez.org/.在4.46上,BlueZ实现了对A2DP Sink的支持,而之前的版本只支持A2DP Source.
- 关于face alglimnment各种资料,存下来有空慢慢看
人脸对齐和应用 机器学习--详解人脸对齐算法SDM-LBF 基于MTCNN的人脸自动对齐技术原理及其Tensorflow实现测试 人脸检测——MTCNN CVPR论文<Face Alignmen ...
- mac上怎么安装dmg
双击dmg文件,就会打开了,里面一般就是应用程序,拖到Finder-应用程序(如果里面是pkg格式,就是安装包,双击安装),然后还要注意一个安装完了以后,要把刚才载入的dmg推出,方法是把桌面上那个图 ...
- flume中HdfsSink参数说明
flume到hdfsSink: type hdfs path 写入hdfs的路径,需要包含文件系统标识,比如:hdfs://namenode/flume/webdata/ 可以使用flume提供的日期 ...
- [zhuan]《MEF程序设计指南》博文汇总
http://www.cnblogs.com/beniao/archive/2010/08/11/1797537.html 在MEF之前,人们已经提出了许多依赖注入框架来解决应用的扩展性问题,比如OS ...
- c数据结构第一个公式写程序计算给定多项式在给定点x处的值
假设x=2,那么a的数组有几个, 那n就是根据索引来的 假设数组[, , , , ] n就是0, 1, 2,,3, 4 f = + ( + ( + ( + ()))) f = + ( + ( + ( ...
- 第三百二十二节,web爬虫,requests请求
第三百二十二节,web爬虫,requests请求 requests请求,就是用yhthon的requests模块模拟浏览器请求,返回html源码 模拟浏览器请求有两种,一种是不需要用户登录或者验证的请 ...
- JDBC事务提交/回滚实例
以下是使用事务教程中描述的提交和回滚的代码示例. 此示例代码是基于前面章节中完成的环境和数据库设置编写的. 复制并将以下示例代码保存到:CommitAndRollback.java 中,编译并运行如下 ...
- (转)YUV420、YUV422、RGB24转换
//平面YUV422转平面RGB24static void YUV422p_to_RGB24(unsigned char *yuv422[3], unsigned char *rgb24, int w ...
- Office在线预览及PDF在线预览的实现方式
原文链接:http://www.officeweb365.com/officetoview.html 一.服务器先转换为PDF,再转换为SWF,最后通过网页加载Flash预览 微软方:利用Office ...
