吴裕雄--天生自然 R语言开发学习:图形初阶(续二)








# ----------------------------------------------------#
# R in Action (2nd ed): Chapter 3 #
# Getting started with graphs #
# requires that the Hmisc and RColorBrewer packages #
# have been installed #
# install.packages(c("Hmisc", "RColorBrewer")) #
#-----------------------------------------------------# par(ask=TRUE)
opar <- par(no.readonly=TRUE) # make a copy of current settings attach(mtcars) # be sure to execute this line plot(wt, mpg)
abline(lm(mpg~wt))
title("Regression of MPG on Weight")
# Input data for drug example
dose <- c(20, 30, 40, 45, 60)
drugA <- c(16, 20, 27, 40, 60)
drugB <- c(15, 18, 25, 31, 40) plot(dose, drugA, type="b") opar <- par(no.readonly=TRUE) # make a copy of current settings
par(lty=2, pch=17) # change line type and symbol
plot(dose, drugA, type="b") # generate a plot
par(opar) # restore the original settings plot(dose, drugA, type="b", lty=3, lwd=3, pch=15, cex=2) # choosing colors
library(RColorBrewer)
n <- 7
mycolors <- brewer.pal(n, "Set1")
barplot(rep(1,n), col=mycolors) n <- 10
mycolors <- rainbow(n)
pie(rep(1, n), labels=mycolors, col=mycolors)
mygrays <- gray(0:n/n)
pie(rep(1, n), labels=mygrays, col=mygrays) # Listing 3.1 - Using graphical parameters to control graph appearance
dose <- c(20, 30, 40, 45, 60)
drugA <- c(16, 20, 27, 40, 60)
drugB <- c(15, 18, 25, 31, 40)
opar <- par(no.readonly=TRUE)
par(pin=c(2, 3))
par(lwd=2, cex=1.5)
par(cex.axis=.75, font.axis=3)
plot(dose, drugA, type="b", pch=19, lty=2, col="red")
plot(dose, drugB, type="b", pch=23, lty=6, col="blue", bg="green")
par(opar) # Adding text, lines, and symbols
plot(dose, drugA, type="b",
col="red", lty=2, pch=2, lwd=2,
main="Clinical Trials for Drug A",
sub="This is hypothetical data",
xlab="Dosage", ylab="Drug Response",
xlim=c(0, 60), ylim=c(0, 70)) # Listing 3.2 - An Example of Custom Axes
x <- c(1:10)
y <- x
z <- 10/x
opar <- par(no.readonly=TRUE)
par(mar=c(5, 4, 4, 8) + 0.1)
plot(x, y, type="b",
pch=21, col="red",
yaxt="n", lty=3, ann=FALSE)
lines(x, z, type="b", pch=22, col="blue", lty=2)
axis(2, at=x, labels=x, col.axis="red", las=2)
axis(4, at=z, labels=round(z, digits=2),
col.axis="blue", las=2, cex.axis=0.7, tck=-.01)
mtext("y=1/x", side=4, line=3, cex.lab=1, las=2, col="blue")
title("An Example of Creative Axes",
xlab="X values",
ylab="Y=X")
par(opar) # Listing 3.3 - Comparing Drug A and Drug B response by dose
dose <- c(20, 30, 40, 45, 60)
drugA <- c(16, 20, 27, 40, 60)
drugB <- c(15, 18, 25, 31, 40)
opar <- par(no.readonly=TRUE)
par(lwd=2, cex=1.5, font.lab=2)
plot(dose, drugA, type="b",
pch=15, lty=1, col="red", ylim=c(0, 60),
main="Drug A vs. Drug B",
xlab="Drug Dosage", ylab="Drug Response")
lines(dose, drugB, type="b",
pch=17, lty=2, col="blue")
abline(h=c(30), lwd=1.5, lty=2, col="gray")
library(Hmisc)
minor.tick(nx=3, ny=3, tick.ratio=0.5)
legend("topleft", inset=.05, title="Drug Type", c("A","B"),
lty=c(1, 2), pch=c(15, 17), col=c("red", "blue"))
par(opar) # Example of labeling points
attach(mtcars)
plot(wt, mpg,
main="Mileage vs. Car Weight",
xlab="Weight", ylab="Mileage",
pch=18, col="blue")
text(wt, mpg,
row.names(mtcars),
cex=0.6, pos=4, col="red")
detach(mtcars) # View font families
opar <- par(no.readonly=TRUE)
par(cex=1.5)
plot(1:7,1:7,type="n")
text(3,3,"Example of default text")
text(4,4,family="mono","Example of mono-spaced text")
text(5,5,family="serif","Example of serif text")
par(opar) # Combining graphs
attach(mtcars)
opar <- par(no.readonly=TRUE)
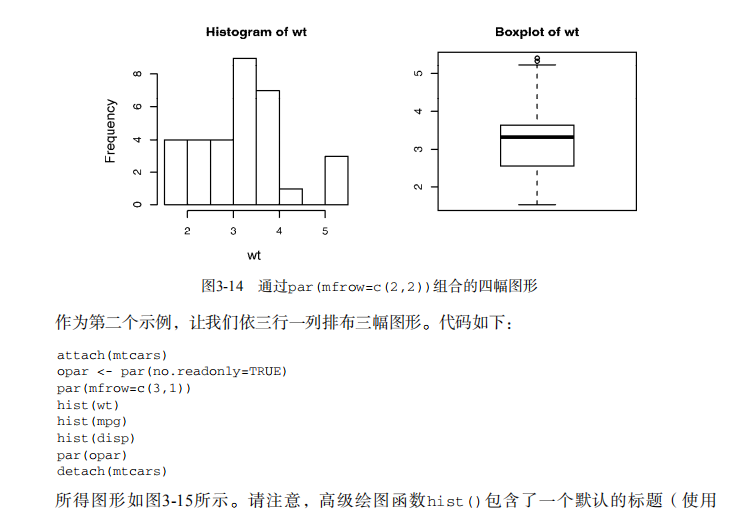
par(mfrow=c(2,2))
plot(wt,mpg, main="Scatterplot of wt vs. mpg")
plot(wt,disp, main="Scatterplot of wt vs. disp")
hist(wt, main="Histogram of wt")
boxplot(wt, main="Boxplot of wt")
par(opar)
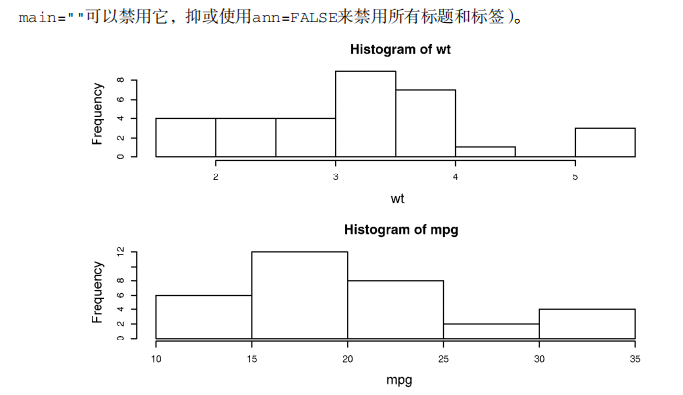
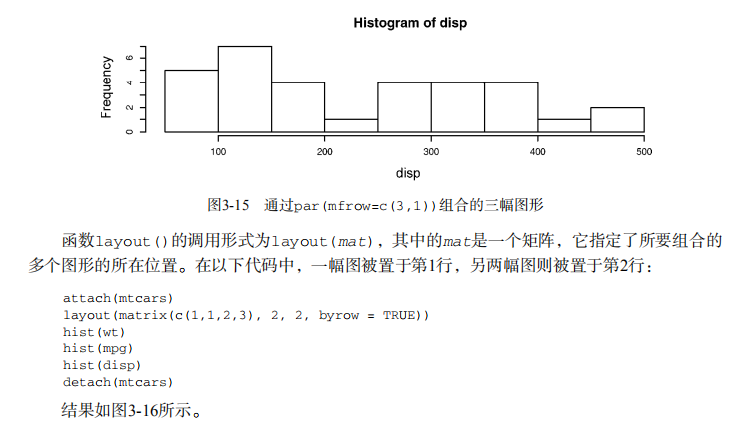
detach(mtcars) attach(mtcars)
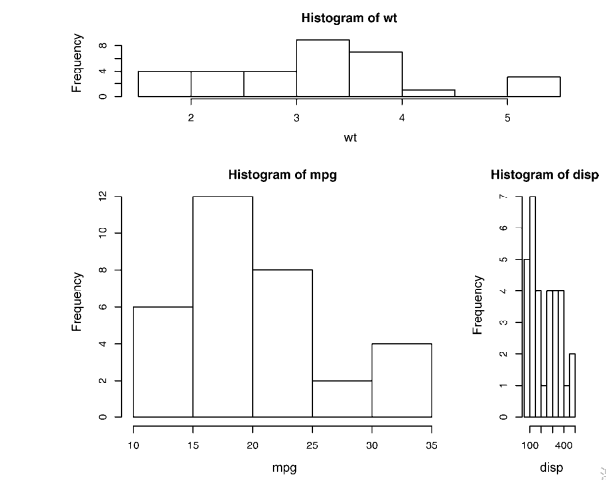
opar <- par(no.readonly=TRUE)
par(mfrow=c(3,1))
hist(wt)
hist(mpg)
hist(disp)
par(opar)
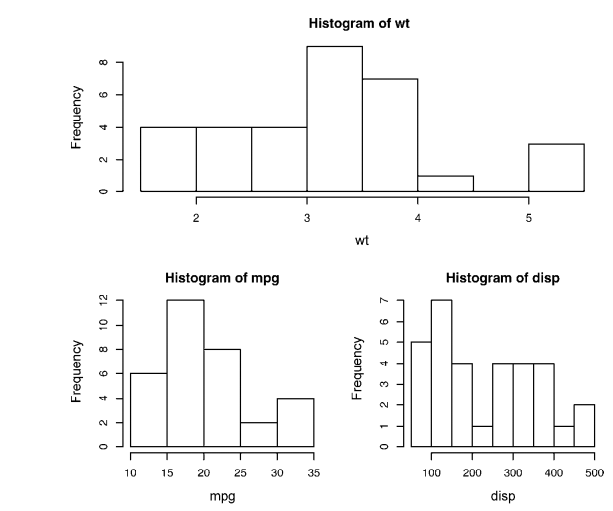
detach(mtcars) attach(mtcars)
layout(matrix(c(1,1,2,3), 2, 2, byrow = TRUE))
hist(wt)
hist(mpg)
hist(disp)
detach(mtcars) attach(mtcars)
layout(matrix(c(1, 1, 2, 3), 2, 2, byrow = TRUE),
widths=c(3, 1), heights=c(1, 2))
hist(wt)
hist(mpg)
hist(disp)
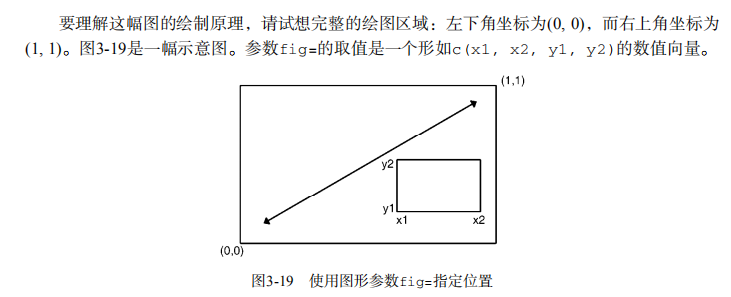
detach(mtcars) # Listing 3.4 - Fine placement of figures in a graph
opar <- par(no.readonly=TRUE)
par(fig=c(0, 0.8, 0, 0.8))
plot(mtcars$mpg, mtcars$wt,
xlab="Miles Per Gallon",
ylab="Car Weight")
par(fig=c(0, 0.8, 0.55, 1), new=TRUE)
boxplot(mtcars$mpg, horizontal=TRUE, axes=FALSE)
par(fig=c(0.65, 1, 0, 0.8), new=TRUE)
boxplot(mtcars$wt, axes=FALSE)
mtext("Enhanced Scatterplot", side=3, outer=TRUE, line=-3)
par(opar)
吴裕雄--天生自然 R语言开发学习:图形初阶(续二)的更多相关文章
- 吴裕雄--天生自然 R语言开发学习:时间序列(续二)
#-----------------------------------------# # R in Action (2nd ed): Chapter 15 # # Time series # # r ...
- 吴裕雄--天生自然 R语言开发学习:方差分析(续二)
#-------------------------------------------------------------------# # R in Action (2nd ed): Chapte ...
- 吴裕雄--天生自然 R语言开发学习:回归(续二)
#------------------------------------------------------------# # R in Action (2nd ed): Chapter 8 # # ...
- 吴裕雄--天生自然 R语言开发学习:分类(续二)
#-----------------------------------------------------------------------------# # R in Action (2nd e ...
- 吴裕雄--天生自然 R语言开发学习:聚类分析(续一)
#-------------------------------------------------------# # R in Action (2nd ed): Chapter 16 # # Clu ...
- 吴裕雄--天生自然 R语言开发学习:时间序列(续三)
#-----------------------------------------# # R in Action (2nd ed): Chapter 15 # # Time series # # r ...
- 吴裕雄--天生自然 R语言开发学习:时间序列(续一)
#-----------------------------------------# # R in Action (2nd ed): Chapter 15 # # Time series # # r ...
- 吴裕雄--天生自然 R语言开发学习:方差分析(续一)
#-------------------------------------------------------------------# # R in Action (2nd ed): Chapte ...
- 吴裕雄--天生自然 R语言开发学习:回归(续四)
#------------------------------------------------------------# # R in Action (2nd ed): Chapter 8 # # ...
- 吴裕雄--天生自然 R语言开发学习:回归(续三)
#------------------------------------------------------------# # R in Action (2nd ed): Chapter 8 # # ...
随机推荐
- IPC---有名管道FIFO
一.参考网址 1.Linux学习之——FIFO实例
- idtcp实现文件下载和上传
unit Unit1; interface uses Windows, Messages, SysUtils, Variants, Classes, Graphics, Controls, Forms ...
- leetcode中的sql
1 组合两张表 组合两张表, 题目很简单, 主要考察JOIN语法的使用.唯一需要注意的一点, 是题目中的这句话, "无论 person 是否有地址信息".说明即使Person表, ...
- Java 14 有哪些新特性?
记录为 Java 提供了一种正确实现数据类的能力,不再需要为实现数据类而编写冗长的代码.下面就来看看 Java 14 中的记录有哪些新特性. 作者 | Nathan Esquenazi 译者 | 弯月 ...
- STL——算法
以下内容大多摘自<C++标准程序库> STL提供了一些标准算法,包括搜寻.排序.拷贝.重新排序.修改.数值运算等.算法并不是容器类别的成员函数,而是一种搭配迭代器使用的全局函数. #inc ...
- Reservoir Computing论文学习
目录 背景: RC优势: 储备池计算主要理论组成: ESNS数学模型 结构表示 状态方程和输出方程 计算过程 储备池的优化 GA:使用进化算法对参数进行优化: 基于随机梯度下降法的储备池参数优化 参考 ...
- 计量经济与时间序列_ACF自相关与PACF偏自相关算法解析(Python,TB(交易开拓者))
1 在时间序列中ACF图和PACF图是非常重要的两个概念,如果运用时间序列做建模.交易或者预测的话.这两个概念是必须的. 2 ACF和PACF分别为:自相关函数(系数)和偏自相关函数(系数). ...
- gcc -S xx
编译器的核心任务是把C程序翻译成机器的汇编语言(assembly language).汇编语言是人类可以阅读的编程语言,也是相当接近实际机器码的语言.由此导致每种 CPU 架构都有不同的汇编语言. 实 ...
- 吴裕雄--天生自然 PYTHON3开发学习:File(文件) 方法
# 打开文件 fo = open("runoob.txt", "wb") print("文件名为: ", fo.name) # 关闭文件 f ...
- 静态、动态cell区别
静态cell:cell数目固定不变,图片/文字固定不变(如qq设置列表可使用静态cell加载) 动态cell:cell数目较多,且图片/文字可能会发生变化(如应网络请求,淘宝列表中某个物品名称或者图片 ...
