uva1368 DNA Consensus String
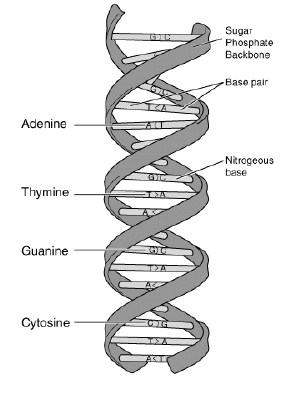 <tex2html_verbatim_mark> Figure 1.
<tex2html_verbatim_mark> Figure 1.DNA (Deoxyribonucleic Acid) is the molecule which contains the genetic instructions. It consists of four different nucleotides, namely Adenine, Thymine, Guanine, and Cytosine as shown in Figure 1. If we represent a nucleotide by its initial character, a DNA strand can be regarded as a long string (sequence of characters) consisting of the four characters A, T, G, and C. For example, assume we are given some part of a DNA strand which is composed of the following sequence of nucleotides:
``Thymine-Adenine-Adenine-Cytosine-Thymine-Guanine-Cytosine-Cytosine-Guanine-Adenine-Thymine"
Then we can represent the above DNA strand with the string ``TAACTGCCGAT." The biologist Prof. Ahn found that a gene X commonly exists in the DNA strands of five different kinds of animals, namely dogs, cats, horses, cows, and monkeys. He also discovered that the DNA sequences of the gene X from each animal were very alike. See Figure 2.
| DNA sequence of gene X | |
| Cat: | GCATATGGCTGTGCA |
| Dog: | GCAAATGGCTGTGCA |
| Horse: | GCTAATGGGTGTCCA |
| Cow: | GCAAATGGCTGTGCA |
| Monkey: | GCAAATCGGTGAGCA |
Prof. Ahn thought that humans might also have the gene X and decided to search for the DNA sequence of X in human DNA. However, before searching, he should define a representative DNA sequence of gene X because its sequences are not exactly the same in the DNA of the five animals. He decided to use the Hamming distance to define the representative sequence. The Hamming distance is the number of different characters at each position from two strings of equal length. For example, assume we are given the two strings ``AGCAT" and ``GGAAT." The Hamming distance of these two strings is 2 because the 1st and the 3rd characters of the two strings are different. Using the Hamming distance, we can define a representative string for a set of multiple strings of equal length. Given a set of strings S = s1,...,sm<tex2html_verbatim_mark> of length n<tex2html_verbatim_mark> , the consensus error between a string y<tex2html_verbatim_mark> of length n<tex2html_verbatim_mark> and the set S<tex2html_verbatim_mark> is the sum of the Hamming distances between y<tex2html_verbatim_mark> and eachsi<tex2html_verbatim_mark> in S<tex2html_verbatim_mark> . If the consensus error between y<tex2html_verbatim_mark> and S<tex2html_verbatim_mark> is the minimum among all possible strings y<tex2html_verbatim_mark> of length n<tex2html_verbatim_mark> , y<tex2html_verbatim_mark> is called a consensus string ofS<tex2html_verbatim_mark> . For example, given the three strings `` AGCAT" `` AGACT" and `` GGAAT" the consensus string of the given strings is `` AGAAT" because the sum of the Hamming distances between `` AGAAT" and the three strings is 3 which is minimal. (In this case, the consensus string is unique, but in general, there can be more than one consensus string.) We use the consensus string as a representative of the DNA sequence. For the example of Figure 2 above, a consensus string of gene X is `` GCAAATGGCTGTGCA" and the consensus error is 7.
Input
Your program is to read from standard input. The input consists of T<tex2html_verbatim_mark> test cases. The number of test cases T<tex2html_verbatim_mark> is given in the first line of the input. Each test case starts with a line containing two integers m<tex2html_verbatim_mark> and n<tex2html_verbatim_mark> which are separated by a single space. The integer m<tex2html_verbatim_mark>(4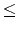 m
m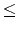 50)<tex2html_verbatim_mark>represents the number of DNA sequences and n<tex2html_verbatim_mark>(4
50)<tex2html_verbatim_mark>represents the number of DNA sequences and n<tex2html_verbatim_mark>(4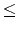 n
n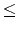 1000)<tex2html_verbatim_mark> represents the length of the DNA sequences, respectively. In each of the next m<tex2html_verbatim_mark> lines, each DNA sequence is given.
1000)<tex2html_verbatim_mark> represents the length of the DNA sequences, respectively. In each of the next m<tex2html_verbatim_mark> lines, each DNA sequence is given.
Output
Your program is to write to standard output. Print the consensus string in the first line of each case and the consensus error in the second line of each case. If there exists more than one consensus string, print the lexicographically smallest consensus string. The following shows sample input and output for three test cases.
Sample Input
3
5 8
TATGATAC
TAAGCTAC
AAAGATCC
TGAGATAC
TAAGATGT
4 10
ACGTACGTAC
CCGTACGTAG
GCGTACGTAT
TCGTACGTAA
6 10
ATGTTACCAT
AAGTTACGAT
AACAAAGCAA
AAGTTACCTT
AAGTTACCAA
TACTTACCAA
Sample Output
TAAGATAC
7
ACGTACGTAA
6
AAGTTACCAA
12
这个问题比较简单,注意字典序就好了。
附上AC代码:
#include<iostream>
#include<algorithm> using namespace std;
char str[][];
char ch[];
int t[];
int n;
int a,b;
int Sum;
int Flag;
int main()
{
cin >> n;
while(n--)
{
Sum=;
cin >> a >> b;
for(int i=;i<a;i++)
cin >> str[i];
for(int i=;i<b;i++)
{
Flag=;
t[]=t[]=t[]=t[]=;
for(int j=;j<a;j++)
{
if(str[j][i]=='A') t[]++;
if(str[j][i]=='C') t[]++;
if(str[j][i]=='G') t[]++;
if(str[j][i]=='T') t[]++;
}
for(int i=;i<;i++)
if(t[i]>t[Flag]) Flag=i;
if(Flag==) cout << 'A';
if(Flag==) cout << 'C';
if(Flag==) cout << 'G';
if(Flag==) cout << 'T';
Sum+=t[Flag];
}
cout << endl;
cout << a*b-Sum << endl;
}
return ;
}
uva1368 DNA Consensus String的更多相关文章
- UVA-1368 DNA Consensus String(思路)
题目: 链接 题意: 题目虽然比较长,但读完之后题目的思路还是比较容易想出来的. 给出m个长度为n的字符串(只包含‘A’.‘T’.‘G’.‘C’),我们的任务是得出一个字符串,要求这个字符串与给出的m ...
- 贪心水题。UVA 11636 Hello World,LA 3602 DNA Consensus String,UVA 10970 Big Chocolate,UVA 10340 All in All,UVA 11039 Building Designing
UVA 11636 Hello World 二的幂答案就是二进制长度减1,不是二的幂答案就是是二进制长度. #include<cstdio> int main() { ; ){ ; ) r ...
- DNA Consensus String
题目(中英对照): DNA (Deoxyribonucleic Acid) is the molecule which contains the genetic instructions. It co ...
- 紫书第三章训练1 E - DNA Consensus String
DNA (Deoxyribonucleic Acid) is the molecule which contains the genetic instructions. It consists of ...
- UVa1368/ZOJ3132 DNA Consensus String
#include <stdio.h>#include <string.h> int main(){ int a[4][1000]; // A/C/G/T在每列中出现的次数 ...
- DNA序列 (DNA Consensus String,ACM/ICPC Seoul 2006,UVa1368
题目描述:算法竞赛入门经典习题3-7 题目思路:每列出现最多的距离即最短 #include <stdio.h> #include <string.h> int main(int ...
- uva 1368 DNA Consensus String
这道题挺简单的,刚开始理解错误,以为是从已有的字符串里面求最短的距离,后面才发现是求一个到所有字符串最小距离的字符串,因为这样的字符串可能有多个,所以最后取最小字典序的字符串. 我的思路就是求每一列每 ...
- uvalive 3602 DNA Consensus String
https://vjudge.net/problem/UVALive-3602 题意: 给定m个长度均为n的DNA序列,求一个DNA序列,使得它到所有的DNA序列的汉明距离最短,若有多个解则输出字典序 ...
- 【每日一题】UVA - 1368 DNA Consensus String 字符串+贪心+阅读题
https://cn.vjudge.net/problem/UVA-1368 二维的hamming距离算法: For binary strings a and b the Hamming distan ...
随机推荐
- repo init 时gpg: 无法检查签名:找不到公钥
i found a solution here: http://www.marshut.com/wrrts/repo-release-1-12-4.html Sorry, I realized tod ...
- an error occured during the file system check
打开虚拟机的时候,报错: 出错原因: 我之前修改了/etc/fstab文件, 原先/etc/fstab文件中有一行是这样的: LABEL=/i01 /u01 ...
- angular+bootstrap分页指令案例
AngularJS中不仅内置了许多指令,而且开发人员也可以通过自定义指令来完成特殊操作,指令创建成功后可以到处复用. Web应用中的分页处理最为常见,我们可以将分页模块编写成一个可以复用的Angula ...
- TVS和一般的稳压二极管有什么区别
电压及电流的瞬态干扰是造成电子电路及设备损坏的主要原因,常给人们带来无法估量的损失.这些干扰通常来自于电力设备的起停操作.交流电网的不稳定.雷击干扰及静电放电等,瞬态干扰几乎无处不在.无时不有,使人感 ...
- Cmake编译成静态库
To build OpenCV as static library you need to set BUILD_SHARED_LIBS flag to false/off: cmake -DBUILD ...
- NOI2011 兔农
http://www.lydsy.com/JudgeOnline/problem.php?id=2432 感觉是day1中最难的一题,还好出题人很良心,给了75分部分分. 还是跪拜策爷吧~Orz ht ...
- TortoiseGit 使用教程
原文地址:http://blog.csdn.net/ethan_xue/article/details/7749639 git的使用越来越广泛 使用命令比较麻烦,下面讲解一下tortoisegit的使 ...
- hdu2769:枚举+同余方程
题意:有一个随机数生成器 x[i+1]=(a*x[i]+b)%10001 已知 x1,x3,x5...求 x2,x4,x6...... x的个数为 2n (n<=10000) a,b也在 0 ...
- Codeforces Round #272 (Div. 1) Problem C. Dreamoon and Strings
C. Dreamoon and Strings time limit per test 1 second memory limit per test 256 megabytes input stand ...
- JDBC中Statement接口提供的execute、executeQuery和executeUpdate之间的区别
Statement 接口提供了三种执行 SQL 语句的方法:executeQuery.executeUpdate 和 execute.使用哪一个方法由 SQL 语句所产生的内容决定. 方法execut ...
