tophat输出结果junction.bed
tophat输出结果junction.bed
| BED format |
|
BED format provides a flexible way to define the data lines that are displayed in an annotation track. BED lines have three required fields and nine additional optional fields. The number of fields per line must be consistent throughout any single set of data in an annotation track. The order of the optional fields is binding: lower-numbered fields must always be populated if higher-numbered fields are used. If your data set is BED-like, but it is very large and you would like to keep it on your own server, you should use the bigBed data format. The first three required BED fields are:
The 9 additional optional BED fields are:
Example:
Example:
Example:
|
tophat输出结果junction.bed中定义的junction就是中间是intron两边是exon的这种序列结构, JUNC00000001包含两个exon和中间的intron
cat junctions.bed
- track name=junctions description="TopHat junctions"
- test_chromosome 177 400 JUNC00000001 61 + 177 400 255,0,0 2 73,50 0,173
- test_chromosome 350 550 JUNC00000002 45 + 350 550 255,0,0 2 50,50 0,150
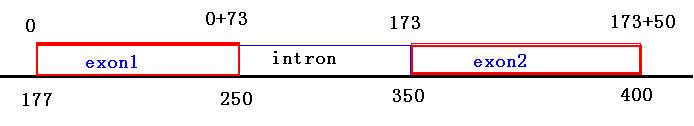
exon1: 177-250(display), 177-249(actual)
intron: 250-350(display), 250-349(actual)
exon2: 350-400(display), 350-399(actual)
chrom(test_chromosome): The name of the chromosome.
这里就是test_chromosome
chromStart(177): The starting position of the feature in the
chromosome or scaffold. The first base in a chromosome is numbered
0.
chromosome上feature(在这里就是junction)的起始位置
chromEnd(400): The ending position of the feature in the chromosome
or scaffold. The chromEnd base is not included in the display of
the feature. For example, the first 100 bases of a chromosome are
defined as chromStart=0, chromEnd=100, and span the bases numbered
0-99.
对应的终止位置
name(JUNC00000001): Defines the name of the BED line.
score(61): A score between 0 and 1000.
strand(+): Defines the strand: either '+' or '-'.
thickStart(177): The starting position at which the feature is
drawn thickly (for example, the start codon in gene
displays).
thickEnd(400): The ending position at which the feature is drawn
thickly (for example, the stop codon in gene displays).
itemRgb(255,0,0): An RGB value of the form R,G,B (e.g. 255,0,0). If
the track line itemRgb attribute is set to "On", this RBG value
will determine the display color of the data contained in this BED
line. NOTE: It is recommended that a simple color scheme (eight
colors or less) be used with this attribute to avoid overwhelming
the color resources of the Genome Browser and your Internet
browser.
blockCount(2): The number of blocks (exons) in the BED line.
外显子个数,2个
blockSizes(73,50): A comma-separated list of the block sizes. The
number of items in this list should correspond to
blockCount.外显子大小,两个数字用逗号隔开
blockStarts(0,173): A comma-separated list of block starts. All of
the blockStart positions should be calculated relative to
chromStart. The number of items in this list should correspond to
blockCount.
外显子起始位置,不从chromosome的起始开始,而是从chromStart开始数,也就是junction的起始位置开始计算,第一个碱基是0
用bed_to_junc转换后得到.juncs其中只保留了junction中intron的部分
cat new_list.juncs
- test_chromosome 249 350 +
- test_chromosome 399 500 +
<chrom> <left> <right> <+/->
http://genome.ucsc.edu/FAQ/FAQformat.html#format1
http://www.ensembl.org/info/website/upload/bed.html
http://zhongguozhanying.blog.163.com/blog/static/21618709520135484450746/
http://blog.sina.com.cn/s/blog_7cffd14001012rtd.html
https://github.com/WormBase/wormbase-pipeline/blob/master/scripts/align_RNASeq.pl
tophat输出结果junction.bed的更多相关文章
- 使用Tophat+cufflinks分析差异表达
使用Tophat+cufflinks分析差异表达 2017-06-15 19:09:43 522 0 0 使用TopHat+Cufflinks的流程图 序列的比对是RNA分析 ...
- RNA-seq连特异性
RNA-seq连特异性 Oct 15, 2015 The strandness of RNA-seq analysis 前段时间一直在研究关于illumina TrueSeq stranded RNA ...
- tophat的用法
概述:tophat是以bowtie2为核心的一款比对软件. tophat工作分两步: 1.将reads用bowtie比对到参考基因组上. 2.将unmapped-reads打断成更小的fragment ...
- TopHat
What is TopHat? TopHat is a program that aligns RNA-Seq reads to a genome in order to identify exon- ...
- RNA-seq差异表达基因分析之TopHat篇
RNA-seq差异表达基因分析之TopHat篇 发表于2012 年 10 月 23 日 TopHat是基于Bowtie的将RNA-Seq数据mapping到参考基因组上,从而鉴定可变剪切(exon-e ...
- poj 1041(字典序输出欧拉回路)
John's trip Time Limit: 1000MS Memory Limit: 65536K Total Submissions: 8641 Accepted: 2893 Spe ...
- 链终止法|边合成边测序|Bowtie|TopHat|Cufflinks|RPKM|FASTX-Toolkit|fastaQC|基因芯片|桥式扩增|
生物信息学 Sanger采用链终止法进行测序 带有荧光基团的ddXTP+其他四种普通的脱氧核苷酸放入同一个培养皿中,例如带有荧光基团的ddATP+普通的脱氧核苷酸A.T.C.G放入同一个培养皿,以此类 ...
- mapreduce多文件输出的两方法
mapreduce多文件输出的两方法 package duogemap; import java.io.IOException; import org.apache.hadoop.conf ...
- Android Studio 多个编译环境配置 多渠道打包 APK输出配置
看完这篇你学到什么: 熟悉gradle的构建配置 熟悉代码构建环境的目录结构,你知道的不仅仅是只有src/main 开发.生成环境等等环境可以任意切换打包 多渠道打包 APK输出文件配置 需求 一般我 ...
随机推荐
- 编译php5.6
没想到编译个LAMP这么麻烦又简单. 按照官网的做就可以了,只是我在CentOs下一直会提示出现这个错误 按照官网的安装方法:install 用下面的参数: ./configure --with-ap ...
- DTCMS展示一级栏目并展示各自栏目下的二级栏目
c#代码中 <!--C#代码--> <%csharp%> string parent_id=DTRequest.GetQueryString("parent_id&q ...
- jquery nicescroll 配置参数
jQuery滚动条插件兼容ie6+.手机.ipad http://www.areaaperta.com/nicescroll/ jQuery(function($){ $("#scrollI ...
- nginx-url重写
location /game_web{ if (!-e $request_filename){//请求不是文件或者目录 rewrite ^/game_web/(\/init/$ last; break ...
- tomcat源码解读(1)–tomcat热部署实现原理
tomcat的热部署实现原理:tomcat启动的时候会有启动一个线程每隔一段时间会去判断应用中加载的类是否发生变法(类总数的变化,类的修改),如果发生了变化就会把应用的启动的线程停止掉,清除引用,并且 ...
- 考研路之C语言
今天在学习C的时候,又学到了一些新的内容,所以赶紧记录下来. case 1: #include <stdio.h> union exa{ struct{ int a; int b; }ou ...
- epoll_create, epoll_ctl和epoll_wait
参考代码 #include <iostream> #include <sys/socket.h> #include <sys/epoll.h> #include & ...
- gulp前端自动化构建工具入门篇
现在我们通过这3个问题来学习一下: 1.什么是gulp? 2.为什么要用gulp? 3.怎么用? 什么是gulp 答:是一个前端自动化的构建工具,直白点说,如果没有这个工具,我们利用人工依旧可以做 ...
- php + mysql + sphinx 的全文检索(2)
简单 使用php api 去查询 sphinx 的索引数据 $sphinx = new SphinxClient(); $sphinx->SetServer ( ...
- 基于Ogre的DeferredShading(延迟渲染)的实现以及应用
http://blog.sina.com.cn/s/blog_458f871201017i06.html 这篇文章写的不错,从比较宏观的角度说了下作者在OGRE中实现的延迟渲染
