Plotting trees from Random Forest models with ggraph
Today, I want to show how I use Thomas Lin Pederson’s awesome ggraph package to plot decision trees from Random Forest models.
I am very much a visual person, so I try to plot as much of my results as possible because it helps me get a better feel for what is going on with my data.
A nice aspect of using tree-based machine learning, like Random Forest models, is that that they are more easily interpreted than e.g. neural networks as they are based on decision trees. So, when I am using such models, I like to plot final decision trees (if they aren’t too large) to get a sense of which decisions are underlying my predictions.
There are a few very convient ways to plot the outcome if you are using the randomForest package but I like to have as much control as possible about the layout, colors, labels, etc. And because I didn’t find a solution I liked for caret models, I developed the following little function (below you may find information about how I built the model):
As input, it takes part of the output from model_rf <- caret::train(... "rf" ...), that gives the trees of the final model: model_rf$finalModel$forest. From these trees, you can specify which one to plot by index.
library(dplyr)
library(ggraph)
library(igraph)
tree_func <- function(final_model,
tree_num) {
# get tree by index
tree <- randomForest::getTree(final_model,
k = tree_num,
labelVar = TRUE) %>%
tibble::rownames_to_column() %>%
# make leaf split points to NA, so the 0s won't get plotted
mutate(`split point` = ifelse(is.na(prediction), `split point`, NA))
# prepare data frame for graph
graph_frame <- data.frame(from = rep(tree$rowname, 2),
to = c(tree$`left daughter`, tree$`right daughter`))
# convert to graph and delete the last node that we don't want to plot
graph <- graph_from_data_frame(graph_frame) %>%
delete_vertices("0")
# set node labels
V(graph)$node_label <- gsub("_", " ", as.character(tree$`split var`))
V(graph)$leaf_label <- as.character(tree$prediction)
V(graph)$split <- as.character(round(tree$`split point`, digits = 2))
# plot
plot <- ggraph(graph, 'dendrogram') +
theme_bw() +
geom_edge_link() +
geom_node_point() +
geom_node_text(aes(label = node_label), na.rm = TRUE, repel = TRUE) +
geom_node_label(aes(label = split), vjust = 2.5, na.rm = TRUE, fill = "white") +
geom_node_label(aes(label = leaf_label, fill = leaf_label), na.rm = TRUE,
repel = TRUE, colour = "white", fontface = "bold", show.legend = FALSE) +
theme(panel.grid.minor = element_blank(),
panel.grid.major = element_blank(),
panel.background = element_blank(),
plot.background = element_rect(fill = "white"),
panel.border = element_blank(),
axis.line = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_blank(),
axis.ticks = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
plot.title = element_text(size = 18))
print(plot)
}We can now plot, e.g. the tree with the smalles number of nodes:
tree_num <- which(model_rf$finalModel$forest$ndbigtree == min(model_rf$finalModel$forest$ndbigtree))
tree_func(final_model = model_rf$finalModel, tree_num)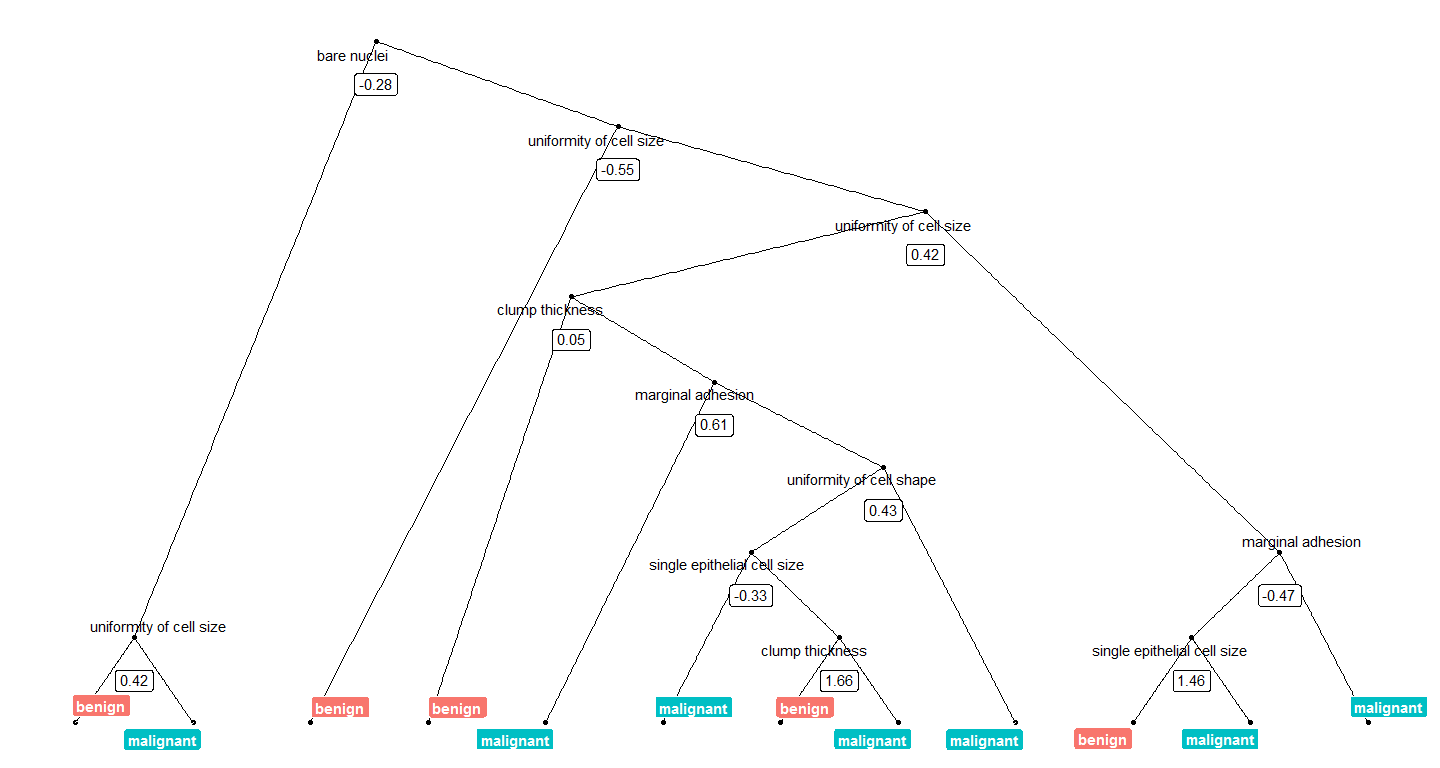
Or we can plot the tree with the biggest number of nodes:
tree_num <- which(model_rf$finalModel$forest$ndbigtree == max(model_rf$finalModel$forest$ndbigtree))
tree_func(final_model = model_rf$finalModel, tree_num)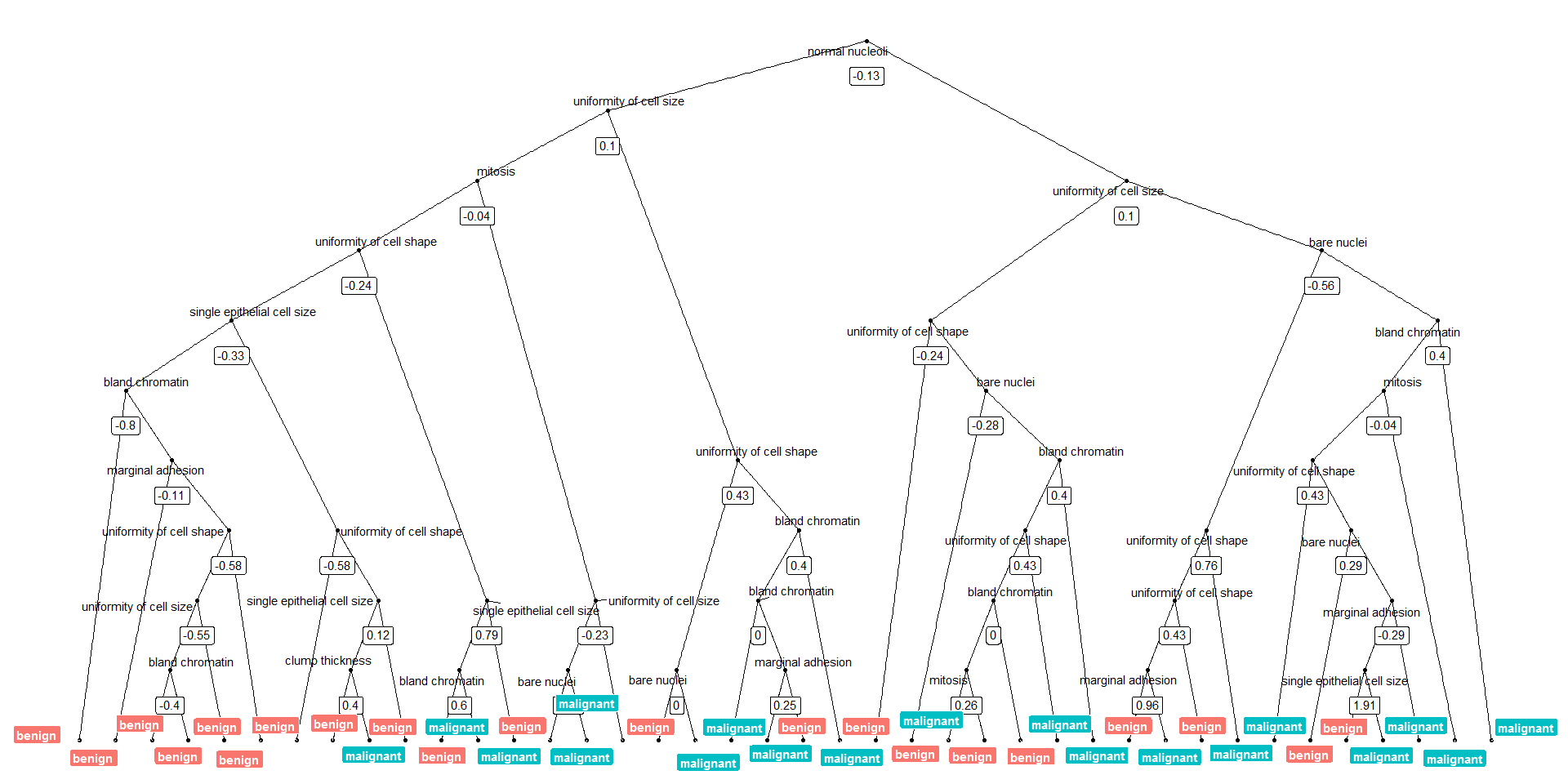
Preparing the data and modeling
The data set I am using in these example analyses, is the Breast Cancer Wisconsin (Diagnostic) Dataset. The data was downloaded from the UC Irvine Machine Learning Repository.
The first data set looks at the predictor classes:
- malignant or
- benign breast mass.
The features characterize cell nucleus properties and were generated from image analysis of fine needle aspirates (FNA) of breast masses:
- Sample ID (code number)
- Clump thickness
- Uniformity of cell size
- Uniformity of cell shape
- Marginal adhesion
- Single epithelial cell size
- Number of bare nuclei
- Bland chromatin
- Number of normal nuclei
- Mitosis
- Classes, i.e. diagnosis
bc_data <- read.table("datasets/breast-cancer-wisconsin.data.txt", header = FALSE, sep = ",")
colnames(bc_data) <- c("sample_code_number",
"clump_thickness",
"uniformity_of_cell_size",
"uniformity_of_cell_shape",
"marginal_adhesion",
"single_epithelial_cell_size",
"bare_nuclei",
"bland_chromatin",
"normal_nucleoli",
"mitosis",
"classes")
bc_data$classes <- ifelse(bc_data$classes == "2", "benign",
ifelse(bc_data$classes == "4", "malignant", NA))
bc_data[bc_data == "?"] <- NA
# impute missing data
library(mice)
bc_data[,2:10] <- apply(bc_data[, 2:10], 2, function(x) as.numeric(as.character(x)))
dataset_impute <- mice(bc_data[, 2:10], print = FALSE)
bc_data <- cbind(bc_data[, 11, drop = FALSE], mice::complete(dataset_impute, 1))
bc_data$classes <- as.factor(bc_data$classes)
# how many benign and malignant cases are there?
summary(bc_data$classes)
# separate into training and test data
library(caret)
set.seed(42)
index <- createDataPartition(bc_data$classes, p = 0.7, list = FALSE)
train_data <- bc_data[index, ]
test_data <- bc_data[-index, ]
# run model
set.seed(42)
model_rf <- caret::train(classes ~ .,
data = train_data,
method = "rf",
preProcess = c("scale", "center"),
trControl = trainControl(method = "repeatedcv",
number = 10,
repeats = 10,
savePredictions = TRUE,
verboseIter = FALSE))If you are interested in more machine learning posts, check out the category listing for machine_learning on my blog.
sessionInfo()## R version 3.3.3 (2017-03-06)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 7 x64 (build 7601) Service Pack 1
##
## locale:
## [1] LC_COLLATE=English_United States.1252
## [2] LC_CTYPE=English_United States.1252
## [3] LC_MONETARY=English_United States.1252
## [4] LC_NUMERIC=C
## [5] LC_TIME=English_United States.1252
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] igraph_1.0.1 ggraph_1.0.0 ggplot2_2.2.1.9000
## [4] dplyr_0.5.0
##
## loaded via a namespace (and not attached):
## [1] Rcpp_0.12.9 nloptr_1.0.4 plyr_1.8.4
## [4] viridis_0.3.4 iterators_1.0.8 tools_3.3.3
## [7] digest_0.6.12 lme4_1.1-12 evaluate_0.10
## [10] tibble_1.2 gtable_0.2.0 nlme_3.1-131
## [13] lattice_0.20-34 mgcv_1.8-17 Matrix_1.2-8
## [16] foreach_1.4.3 DBI_0.6 ggrepel_0.6.5
## [19] yaml_2.1.14 parallel_3.3.3 SparseM_1.76
## [22] gridExtra_2.2.1 stringr_1.2.0 knitr_1.15.1
## [25] MatrixModels_0.4-1 stats4_3.3.3 rprojroot_1.2
## [28] grid_3.3.3 caret_6.0-73 nnet_7.3-12
## [31] R6_2.2.0 rmarkdown_1.3 minqa_1.2.4
## [34] udunits2_0.13 tweenr_0.1.5 deldir_0.1-12
## [37] reshape2_1.4.2 car_2.1-4 magrittr_1.5
## [40] units_0.4-2 backports_1.0.5 scales_0.4.1
## [43] codetools_0.2-15 ModelMetrics_1.1.0 htmltools_0.3.5
## [46] MASS_7.3-45 splines_3.3.3 randomForest_4.6-12
## [49] assertthat_0.1 pbkrtest_0.4-6 ggforce_0.1.1
## [52] colorspace_1.3-2 labeling_0.3 quantreg_5.29
## [55] stringi_1.1.2 lazyeval_0.2.0 munsell_0.4.3转自:https://shiring.github.io/machine_learning/2017/03/16/rf_plot_ggraph
Plotting trees from Random Forest models with ggraph的更多相关文章
- 机器学习算法 --- Pruning (decision trees) & Random Forest Algorithm
一.Table for Content 在之前的文章中我们介绍了Decision Trees Agorithms,然而这个学习算法有一个很大的弊端,就是很容易出现Overfitting,为了解决此问题 ...
- Random Forest And Extra Trees
随机森林 我们对使用决策树随机取样的集成学习有个形象的名字–随机森林. scikit-learn 中封装的随机森林,在决策树的节点划分上,在随机的特征子集上寻找最优划分特征. import numpy ...
- Random Forest Classification of Mushrooms
There is a plethora of classification algorithms available to people who have a bit of coding experi ...
- 机器学习方法(六):随机森林Random Forest,bagging
欢迎转载,转载请注明:本文出自Bin的专栏blog.csdn.net/xbinworld. 技术交流QQ群:433250724,欢迎对算法.技术感兴趣的同学加入. 前面机器学习方法(四)决策树讲了经典 ...
- [Machine Learning & Algorithm] 随机森林(Random Forest)
1 什么是随机森林? 作为新兴起的.高度灵活的一种机器学习算法,随机森林(Random Forest,简称RF)拥有广泛的应用前景,从市场营销到医疗保健保险,既可以用来做市场营销模拟的建模,统计客户来 ...
- paper 85:机器统计学习方法——CART, Bagging, Random Forest, Boosting
本文从统计学角度讲解了CART(Classification And Regression Tree), Bagging(bootstrap aggregation), Random Forest B ...
- 多分类问题中,实现不同分类区域颜色填充的MATLAB代码(demo:Random Forest)
之前建立了一个SVM-based Ordinal regression模型,一种特殊的多分类模型,就想通过可视化的方式展示模型分类的效果,对各个分类区域用不同颜色表示.可是,也看了很多代码,但基本都是 ...
- 统计学习方法——CART, Bagging, Random Forest, Boosting
本文从统计学角度讲解了CART(Classification And Regression Tree), Bagging(bootstrap aggregation), Random Forest B ...
- sklearn_随机森林random forest原理_乳腺癌分类器建模(推荐AAA)
sklearn实战-乳腺癌细胞数据挖掘(博主亲自录制视频) https://study.163.com/course/introduction.htm?courseId=1005269003& ...
随机推荐
- Java --- JSP2新特性
自从03年发布了jsp2.0之后,新增了一些额外的特性,这些特性使得动态网页设计变得更加容易.jsp2.0以后的版本统称jsp2.主要的新增特性有如下几个: 直接配置jsp属性 表达式语言(EL) 标 ...
- HTTP协议介绍
一.什么是HTTP协议呢? 维基百科 写道 超文本传输协议(英文:HyperText Transfer Protocol,缩写:HTTP)是互联网上应用最为广泛的一种网络协议.HTTP是一个客户端终端 ...
- java多线程基本概述(二十)——中断
线程中断我们已经直到可以使用 interrupt() 方法,但是你必须要持有 Thread 对象,但是新的并发库中似乎在避免直接对 Thread 对象的直接操作,尽量使用 Executor 来执行所有 ...
- js字符串的操作
js中字符串的使用非常普遍,以下是一些常用的方法和属性,字符串以str='abcdabc'举例. 1.length属性 获取字符串的长度,str.length返回7 2.replace()方法 str ...
- CF #Manthan, Codefest 16 C. Spy Syndrome 2 Trie
题目链接:http://codeforces.com/problemset/problem/633/C 大意就是给个字典和一个字符串,求一个用字典中的单词恰好构成字符串的匹配. 比赛的时候是用AC自动 ...
- 《HelloGitHub》第 13 期
公告 本期推荐的项目到达了 30 个,里面少不了对本项目支持的小伙伴们,再次感谢大家. 本次排版尝试:根据分类项目名排序,为了让大家方便查阅.如果有任何建议和意见欢迎留言讨论 临近 5.1 假期,所以 ...
- bootstrap快速入门笔记(九)-响应式工具
一,可用的类 超小屏幕手机 (<768px) 小屏幕平板 (≥768px) 中等屏幕桌面 (≥992px) 大屏幕桌面 (≥1200px) .visible-xs-* 可见 隐藏 隐藏 隐藏 ...
- day01课程回顾,数据类型
Day01 Python的分类 Cpython:代码àc字节码->机器码 一行一行的编译执行 Pypy: 代码àc字节码->机器码 全部转换完再执行 其他python 代码- ...
- 「七天自制PHP框架」第三天:PHP实现的设计模式
往期回顾:「七天自制PHP框架」第二天:模型与数据库,点击此处 原文地址:http://www.cnblogs.com/sweng/p/6624845.html,欢迎关注:编程老头 为什么要使用设计模 ...
- 关于爬楼梯的lintcode代码
讲真的,这个我只会用递归去做,但是lintcode上面超时,所以只有在网上找了个动态规划的,虽然这个程序懂了,但是我觉得还是挺不容易的真正弄懂的话-- class Solution {public: ...
