Best packages for data manipulation in R
dplyr and data.table are amazing packages that make data manipulation in R fun. Both packages have their strengths. While dplyr is more elegant and resembles natural language, data.table is succinct and we can do a lot withdata.table in just a single line. Further, data.table is, in some cases, faster (see benchmark here) and it may be a go-to package when performance and memory are constraints. You can read comparison of dplyr and data.tablefrom Stack Overflow and Quora.
You can get reference manual and vignettes for data.table here and for dplyrhere. You can read other tutorial about dplyr published at DataScience+
Background
I am a long time dplyr and data.table user for my data manipulation tasks. For someone who knows one of these packages, I thought it could help to show codes that perform the same tasks in both packages to help them quickly study the other. If you know either package and have interest to study the other, this post is for you.
dplyr
dplyr has 5 verbs which make up the majority of the data manipulation tasks we perform. Select: used to select one or more columns; Filter: used to select some rows based on specific criteria; Arrange: used to sort data based on one or more columns in ascending or descending order; Mutate: used to add new columns to our data; Summarise: used to create chunks from our data.
data.table
data.table has a very succinct general format: DT[i, j, by], which is interpreted as: Take DT, subset rows using i, then calculate j grouped by by.
Data manipulation
First we will install some packages for our project.
- library(dplyr)
- library(data.table)
- library(lubridate)
- library(jsonlite)
- library(tidyr)
- library(ggplot2)
- library(compare)
The data we will use here is from DATA.GOV. It is Medicare Hospital Spending by Claim and it can be downloaded from here. Let’s download the data in JSONformat using the fromJSON function from the jsonlite package. Since JSON is a very common data format used for asynchronous browser/server communication, it is good if you understand the lines of code below used to get the data. You can get an introductory tutorial on how to use the jsonlite package to work with JSON data here and here. However, if you want to focus only on the data.table and dplyr commands, you can safely just run the codes in the two cells below and ignore the details.
- spending=fromJSON("https://data.medicare.gov/api/views/nrth-mfg3/rows.json?accessType=DOWNLOAD")
- names(spending)
- "meta" "data"
- meta=spending$meta
- hospital_spending=data.frame(spending$data)
- colnames(hospital_spending)=make.names(meta$view$columns$name)
- hospital_spending=select(hospital_spending,-c(sid:meta))
- glimpse(hospital_spending)
- Observations: 70598
- Variables:
- $ Hospital.Name (fctr) SOUTHEAST ALABAMA MEDICAL CENT...
- $ Provider.Number. (fctr) 010001, 010001, 010001, 010001...
- $ State (fctr) AL, AL, AL, AL, AL, AL, AL, AL...
- $ Period (fctr) 1 to 3 days Prior to Index Hos...
- $ Claim.Type (fctr) Home Health Agency, Hospice, I...
- $ Avg.Spending.Per.Episode..Hospital. (fctr) 12, 1, 6, 160, 1, 6, 462, 0, 0...
- $ Avg.Spending.Per.Episode..State. (fctr) 14, 1, 6, 85, 2, 9, 492, 0, 0,...
- $ Avg.Spending.Per.Episode..Nation. (fctr) 13, 1, 5, 117, 2, 9, 532, 0, 0...
- $ Percent.of.Spending..Hospital. (fctr) 0.06, 0.01, 0.03, 0.84, 0.01, ...
- $ Percent.of.Spending..State. (fctr) 0.07, 0.01, 0.03, 0.46, 0.01, ...
- $ Percent.of.Spending..Nation. (fctr) 0.07, 0.00, 0.03, 0.58, 0.01, ...
- $ Measure.Start.Date (fctr) 2014-01-01T00:00:00, 2014-01-0...
- $ Measure.End.Date (fctr) 2014-12-31T00:00:00, 2014-12-3...
As shown above, all columns are imported as factors and let’s change the columns that contain numeric values to numeric.
- cols = 6:11; # These are the columns to be changed to numeric.
- hospital_spending[,cols] <- lapply(hospital_spending[,cols], as.numeric)
The last two columns are measure start date and measure end date. So, let’s use the lubridate package to correct the classes of these columns.
- cols = 12:13; # These are the columns to be changed to dates.
- hospital_spending[,cols] <- lapply(hospital_spending[,cols], ymd_hms)
Now, let’s check if the columns have the classes we want.
- sapply(hospital_spending, class)
- $Hospital.Name
- "factor"
- $Provider.Number.
- "factor"
- $State
- "factor"
- $Period
- "factor"
- $Claim.Type
- "factor"
- $Avg.Spending.Per.Episode..Hospital.
- "numeric"
- $Avg.Spending.Per.Episode..State.
- "numeric"
- $Avg.Spending.Per.Episode..Nation.
- "numeric"
- $Percent.of.Spending..Hospital.
- "numeric"
- $Percent.of.Spending..State.
- "numeric"
- $Percent.of.Spending..Nation.
- "numeric"
- $Measure.Start.Date
- "POSIXct" "POSIXt"
- $Measure.End.Date
- "POSIXct" "POSIXt"
Create data table
We can create a data.table using the data.table() function.
- hospital_spending_DT = data.table(hospital_spending)
- class(hospital_spending_DT)
- "data.table" "data.frame"
Select certain columns of data
To select columns, we use the verb select in dplyr. In data.table, on the other hand, we can specify the column names.
Selecting one variable
Let’s selet the “Hospital Name” variable
- from_dplyr = select(hospital_spending, Hospital.Name)
- from_data_table = hospital_spending_DT[,.(Hospital.Name)]
Now, let’s compare if the results from dplyr and data.table are the same.
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- dropped attributes
Removing one variable
- from_dplyr = select(hospital_spending, -Hospital.Name)
- from_data_table = hospital_spending_DT[,!c("Hospital.Name"),with=FALSE]
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- dropped attributes
we can also use := function which modifies the input data.table by reference.
We will use the copy() function, which deep copies the input object and therefore any subsequent update by reference operations performed on the copied object will not affect the original object.
- DT=copy(hospital_spending_DT)
- DT=DT[,Hospital.Name:=NULL]
- "Hospital.Name"%in%names(DT)FALSE
We can also remove many variables at once similarly:
- DT=copy(hospital_spending_DT)
- DT=DT[,c("Hospital.Name","State","Measure.Start.Date","Measure.End.Date"):=NULL]
- c("Hospital.Name","State","Measure.Start.Date","Measure.End.Date")%in%names(DT)
- FALSE FALSE FALSE FALSE
Selecting multiple variables
Let’s select the variables:
Hospital.Name,State,Measure.Start.Date,and Measure.End.Date.
- from_dplyr = select(hospital_spending, Hospital.Name,State,Measure.Start.Date,Measure.End.Date)
- from_data_table = hospital_spending_DT[,.(Hospital.Name,State,Measure.Start.Date,Measure.End.Date)]
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- dropped attributes
Dropping multiple variables
Now, let’s remove the variables Hospital.Name,State,Measure.Start.Date,and Measure.End.Date from the original data frame hospital_spending and the data.table hospital_spending_DT.
- from_dplyr = select(hospital_spending, -c(Hospital.Name,State,Measure.Start.Date,Measure.End.Date))
- from_data_table = hospital_spending_DT[,!c("Hospital.Name","State","Measure.Start.Date","Measure.End.Date"),with=FALSE]
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- dropped attributes
dplyr has functions contains(), starts_with() and, ends_with() which we can use with the verb select. In data.table, we can use regular expressions. Let’s select columns that contain the word Date to demonstrate by example.
- from_dplyr = select(hospital_spending,contains("Date"))
- from_data_table = subset(hospital_spending_DT,select=grep("Date",names(hospital_spending_DT)))
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- dropped attributes
- names(from_dplyr)
- "Measure.Start.Date" "Measure.End.Date"
Rename columns
- setnames(hospital_spending_DT,c("Hospital.Name", "Measure.Start.Date","Measure.End.Date"), c("Hospital","Start_Date","End_Date"))
- names(hospital_spending_DT)
- "Hospital" "Provider.Number." "State" "Period" "Claim.Type" "Avg.Spending.Per.Episode..Hospital." "Avg.Spending.Per.Episode..State." "Avg.Spending.Per.Episode..Nation." "Percent.of.Spending..Hospital." "Percent.of.Spending..State." "Percent.of.Spending..Nation." "Start_Date" "End_Date"
- hospital_spending = rename(hospital_spending,Hospital= Hospital.Name, Start_Date=Measure.Start.Date,End_Date=Measure.End.Date)
- compare(hospital_spending,hospital_spending_DT, allowAll=TRUE)
- TRUE
- dropped attributes
Filtering data to select certain rows
To filter data to select specific rows, we use the verb filter from dplyr with logical statements that could include regular expressions. In data.table, we need the logical statements only.
Filter based on one variable
- from_dplyr = filter(hospital_spending,State=='CA') # selecting rows for California
- from_data_table = hospital_spending_DT[State=='CA']
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- dropped attributes
Filter based on multiple variables
- from_dplyr = filter(hospital_spending,State=='CA' & Claim.Type!="Hospice")
- from_data_table = hospital_spending_DT[State=='CA' & Claim.Type!="Hospice"]
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- dropped attributes
- from_dplyr = filter(hospital_spending,State %in% c('CA','MA',"TX"))
- from_data_table = hospital_spending_DT[State %in% c('CA','MA',"TX")]
- unique(from_dplyr$State)
- CA MA TX
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- dropped attributes
Order data
We use the verb arrange in dplyr to order the rows of data. We can order the rows by one or more variables. If we want descending, we have to use desc()as shown in the examples.The examples are self-explanatory on how to sort in ascending and descending order. Let’s sort using one variable.
Ascending
- from_dplyr = arrange(hospital_spending, State)
- from_data_table = setorder(hospital_spending_DT, State)
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- dropped attributes
Descending
- from_dplyr = arrange(hospital_spending, desc(State))
- from_data_table = setorder(hospital_spending_DT, -State)
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- dropped attributes
Sorting with multiple variables
Let’s sort with State in ascending order and End_Date in descending order.
- from_dplyr = arrange(hospital_spending, State,desc(End_Date))
- from_data_table = setorder(hospital_spending_DT, State,-End_Date)
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- dropped attributes
Adding/updating column(s)
In dplyr we use the function mutate() to add columns. In data.table, we can Add/update a column by reference using := in one line.
- from_dplyr = mutate(hospital_spending, diff=Avg.Spending.Per.Episode..State. - Avg.Spending.Per.Episode..Nation.)
- from_data_table = copy(hospital_spending_DT)
- from_data_table = from_data_table[,diff := Avg.Spending.Per.Episode..State. - Avg.Spending.Per.Episode..Nation.]
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- sorted
- renamed rows
- dropped row names
- dropped attributes
- from_dplyr = mutate(hospital_spending, diff1=Avg.Spending.Per.Episode..State. - Avg.Spending.Per.Episode..Nation.,diff2=End_Date-Start_Date)
- from_data_table = copy(hospital_spending_DT)
- from_data_table = from_data_table[,c("diff1","diff2") := list(Avg.Spending.Per.Episode..State. - Avg.Spending.Per.Episode..Nation.,diff2=End_Date-Start_Date)]
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- dropped attributes
Summarizing columns
We can use the summarize() function from dplyr to create summary statistics.
- summarize(hospital_spending,mean=mean(Avg.Spending.Per.Episode..Nation.))
- mean 8.772727
- hospital_spending_DT[,.(mean=mean(Avg.Spending.Per.Episode..Nation.))]
- mean 8.772727
- summarize(hospital_spending,mean=mean(Avg.Spending.Per.Episode..Nation.),
- maximum=max(Avg.Spending.Per.Episode..Nation.),
- minimum=min(Avg.Spending.Per.Episode..Nation.),
- median=median(Avg.Spending.Per.Episode..Nation.))
- mean maximum minimum median
- 8.77 19 1 8.5
- hospital_spending_DT[,.(mean=mean(Avg.Spending.Per.Episode..Nation.),
- maximum=max(Avg.Spending.Per.Episode..Nation.),
- minimum=min(Avg.Spending.Per.Episode..Nation.),
- median=median(Avg.Spending.Per.Episode..Nation.))]
- mean maximum minimum median
- 8.77 19 1 8.5
We can calculate our summary statistics for some chunks separately. We use the function group_by() in dplyr and in data.table, we simply provide by.
- head(hospital_spending_DT[,.(mean=mean(Avg.Spending.Per.Episode..Hospital.)),by=.(Hospital)])
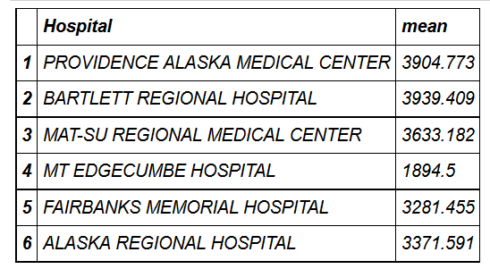
- mygroup= group_by(hospital_spending,Hospital)
- from_dplyr = summarize(mygroup,mean=mean(Avg.Spending.Per.Episode..Hospital.))
- from_data_table=hospital_spending_DT[,.(mean=mean(Avg.Spending.Per.Episode..Hospital.)), by=.(Hospital)]
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- sorted
- renamed rows
- dropped row names
- dropped attributes
We can also provide more than one grouping condition.
- head(hospital_spending_DT[,.(mean=mean(Avg.Spending.Per.Episode..Hospital.)),
- by=.(Hospital,State)])
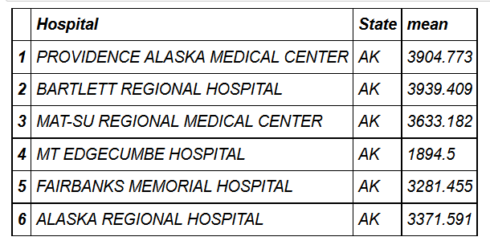
- mygroup= group_by(hospital_spending,Hospital,State)
- from_dplyr = summarize(mygroup,mean=mean(Avg.Spending.Per.Episode..Hospital.))
- from_data_table=hospital_spending_DT[,.(mean=mean(Avg.Spending.Per.Episode..Hospital.)), by=.(Hospital,State)]
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- sorted
- renamed rows
- dropped row names
- dropped attributes
Chaining
With both dplyr and data.table, we can chain functions in succession. In dplyr, we use pipes from the magrittr package with %>% which is really cool. %>% takes the output from one function and feeds it to the first argument of the next function. In data.table, we can use %>% or [ for chaining.
- from_dplyr=hospital_spending%>%group_by(Hospital,State)%>%summarize(mean=mean(Avg.Spending.Per.Episode..Hospital.))
- from_data_table=hospital_spending_DT[,.(mean=mean(Avg.Spending.Per.Episode..Hospital.)), by=.(Hospital,State)]
- compare(from_dplyr,from_data_table, allowAll=TRUE)
- TRUE
- sorted
- renamed rows
- dropped row names
- dropped attributes
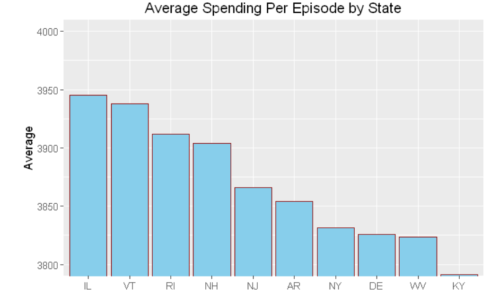
- hospital_spending%>%group_by(State)%>%summarize(mean=mean(Avg.Spending.Per.Episode..Hospital.))%>%
- arrange(desc(mean))%>%head(10)%>%
- mutate(State = factor(State,levels = State[order(mean,decreasing =TRUE)]))%>%
- ggplot(aes(x=State,y=mean))+geom_bar(stat='identity',color='darkred',fill='skyblue')+
- xlab("")+ggtitle('Average Spending Per Episode by State')+
- ylab('Average')+ coord_cartesian(ylim = c(3800, 4000))
- hospital_spending_DT[,.(mean=mean(Avg.Spending.Per.Episode..Hospital.)),
- by=.(State)][order(-mean)][1:10]%>%
- mutate(State = factor(State,levels = State[order(mean,decreasing =TRUE)]))%>%
- ggplot(aes(x=State,y=mean))+geom_bar(stat='identity',color='darkred',fill='skyblue')+
- xlab("")+ggtitle('Average Spending Per Episode by State')+
- ylab('Average')+ coord_cartesian(ylim = c(3800, 4000))

Summary
In this blog post, we saw how we can perform the same tasks using data.tableand dplyr packages. Both packages have their strengths. While dplyr is more elegant and resembles natural language, data.table is succinct and we can do a lot with data.table in just a single line. Further, data.table is, in some cases, faster and it may be a go-to package when performance and memory are the constraints.
You can get the code for this blog post at my GitHub account.
This is enough for this post. If you have any questions or feedback, feel free to leave a comment.
转自:http://datascienceplus.com/best-packages-for-data-manipulation-in-r/
Best packages for data manipulation in R的更多相关文章
- Data manipulation primitives in R and Python
Data manipulation primitives in R and Python Both R and Python are incredibly good tools to manipula ...
- Data Manipulation with dplyr in R
目录 select The filter and arrange verbs arrange filter Filtering and arranging Mutate The count verb ...
- The dplyr package has been updated with new data manipulation commands for filters, joins and set operations.(转)
dplyr 0.4.0 January 9, 2015 in Uncategorized I’m very pleased to announce that dplyr 0.4.0 is now av ...
- An Introduction to Stock Market Data Analysis with R (Part 1)
Around September of 2016 I wrote two articles on using Python for accessing, visualizing, and evalua ...
- 7 Tools for Data Visualization in R, Python, and Julia
7 Tools for Data Visualization in R, Python, and Julia Last week, some examples of creating visualiz ...
- java.sql.SQLException: Can not issue data manipulation statements with executeQuery().
1.错误描写叙述 java.sql.SQLException: Can not issue data manipulation statements with executeQuery(). at c ...
- Can not issue data manipulation statements with executeQuery()错误解决
转: Can not issue data manipulation statements with executeQuery()错误解决 2012年03月27日 15:47:52 katalya 阅 ...
- 数据库原理及应用-SQL数据操纵语言(Data Manipulation Language)和嵌入式SQL&存储过程
2018-02-19 18:03:54 一.数据操纵语言(Data Manipulation Language) 数据操纵语言是指插入,删除和更新语言. 二.视图(View) 数据库三级模式,两级映射 ...
- Can not issue data manipulation statements with executeQuery().解决方案
这个错误提示是说无法发行sql语句到指定的位置 错误写法: 正确写法: excuteQuery是查询语句,而我要调用的是更新的语句,所以这样数据库很为难到底要干嘛,实际我想用的是更新,但是我写成了查询 ...
随机推荐
- Android中实现定时器的四种方式
第一种方式利用Timer和TimerTask 1.继承关系 java.util.Timer 基本方法 schedule 例如: timer.schedule(task, delay,period); ...
- Webdriver API之操作(二)
一.窗口截图 dirver.get_screenshot_as_file("D:\\report\\image\\xxx.jpg") 二.关闭窗口 dirver.close() # ...
- Ajax 与 Comet
Ajax技术的核心是XMLHttpRequest对象(简称XHR). XMLHttpRequest对象 在浏览器中创建XHR对象要像下面这样,使用XMLHttpRequest构造函数. var xhr ...
- JS绑定种类汇总
这里是<你不知道的JS>中常见的this绑定种类分享: 1)默认绑定: function foo(){ console.log(this.a); } var a = 2; foo(); 解 ...
- 使用Eclipse Memory Analyzer Tool(MAT)分析线上故障(一) - 视图&功能篇
Eclipse Memory Analyzer Tool(MAT)相关文章目录: 使用Eclipse Memory Analyzer Tool(MAT)分析线上故障(一) - 视图&功能篇 使 ...
- C#网络程序设计(3)网络传输编程之TCP编程
网络传输编程指基于各种网络协议进行编程,包括TCP编程,UDP编程,P2P编程.本节介绍TCP编程. (1)TCP简介: TCP是TCP/IP体系中最重要的传输层协议,它提供全双工和可 ...
- js表白心形特效
好久没有仔细钻研技术了,闲下来借鉴一下做出一些效果 友情链接: http://tiepeng.applinzi.com/love_you/ ;;background:#ffe;font-size:12 ...
- webService基础知识--认识WebService
之前在找工作的时候,有面试官问到WebService,当时没有接触过,正好现在做的项目中有用到WebService,所以就趁着业余时间来学习了. 一.简介 先来看看百度百科对WebService的解释 ...
- [ext4]03 磁盘布局 – Flexible group分析
Flexible Block Groups (flex_bg),我称之为"弹性块组",是EXT4文件系统引入的一个feature. 所谓Flexible Block Groups, ...
- 蓝桥杯-土地测量-java
/* (程序头部注释开始) * 程序的版权和版本声明部分 * Copyright (c) 2016, 广州科技贸易职业学院信息工程系学生 * All rights reserved. * 文件名称: ...
