【转载】 NeuroEvolution with MarI/O —— 使用人工智能来通关超级玛丽
原文地址:
http://glenn-roberts.com/posts/tech/2015/07/08/neuroevolution-with-mario.html
参考:
https://v.qq.com/x/page/e0532hfg6rp.html
https://www.sohu.com/a/161598493_633698
https://www.jianshu.com/p/7ac0e2bba37c
==================================================
I was recently intrigued by Seth Bling’s MarI/O - a neural network slash genetic algorithm that teaches itself to play Super Mario World.
Seth’s implementation (in Lua) is based on the concept of NeuroEvolution of Augmenting Topologies (or NEAT). NEAT is a type of genetic algorithm which generates efficient artificial neural networks (ANNs) from a very simple starting network. It does so rather quickly too (compared to other evolutionary algorithms).
For another example of why this field is incredibly exciting, watch this amazing video of Google’s DeepMind learning and mastering space invaders. How good is that clutch shot at the end?!
Seth’s MarI/O can play both Super Mario World (SNES), and Super Mario Bros (NES). If you want to try it out yourself, read on.
Setup (Windows 8.1)
To evolve your own ANN with MarI/O that can play Super Mario World, here’s how to do it;
Installation
Install BizHawk Prereqs
Download and unzip BizHawk
Get a copy of Seth’s MarI/O (call it neatevolve.lua )
Put neatevolve.lua in the root folder of your BizHawk folder. (In the same dir as the EmuHawk executable.)
Emulator Setup
Set BizHawk video Mode to OpenGL (not GDI+)
Config > Display > Display Method > Open GL
Restart BizHawk for settings to take effect. Double check it actually works.
Optional: Set emulation speed to 200% - this makes the evolution go a lot faster!
Initial State Setup
We need an initial/fresh game state that gets loaded for each genome. In other words, we need to save the ROM state at the start of the desired level we want MarI/O to learn.
Load the Super Mario World (USA).sfc ROM.
Start a new game
Go to the level you want MarI/O to learn. I chose Yoshi’s Island #1.

- Use the File -> Save Named State -> Save As “DP1.state” in the BizHawk root folder (i.e. in the same dir as neatevolve.lua).
Now we have an initial state that MarI/O will load before each genome is evaluated.
Running MarI/O
Load neatevolve.lua. You can do this via Tools->Lua Console. I prefer to drag and drop neatevolve.lua into the running emulator.
MarI/O will load, creating a base set of about 300 very simple genomes. This is as per the NEAT methodology, which starts with a very simple ANNs (i.e. very few hidden nodes), and evolves from there.
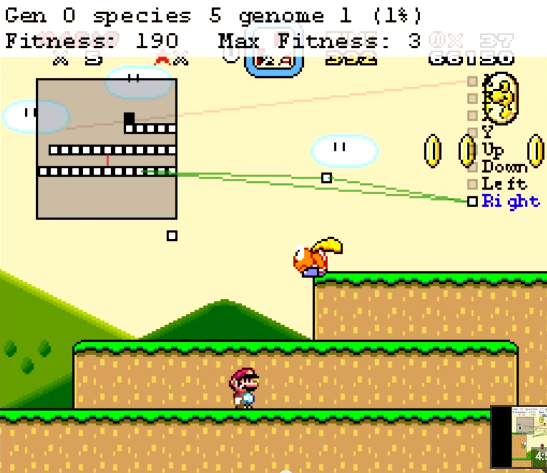
You can see the ANN that MarI/O is currently evaluating by checking ‘Show Map’ setting in the MarI/O ‘Fitness’ window.
Congratulations! If all goes well you’ll see Mario sitting there or jumping up and down, like an idiot, while it learns how to play the game. Don’t worry, it gets ‘smarter’.
Restarting MarI/O
MarI/O saves the genomes of a given generation in a .pool file. The current generation being evaluated is saved in temp.pool. After each generation, a new .pool file will be saved, prefixed with the generation number.
If your computer melts, and you need to restart MarI/O;
- Delete temp.pool
- Copy the desired generation .pool file to DP1.state.pool
- In the MarI/O ‘Fitness’ window, load the DP1.state.pool
- MarI/O should resume from the latest complete generation.
Troubleshooting
Here are solutions to common errors myself an other people have ran into with MarI/O.
‘Buttonnames’ error
LuaInterface.LuaScriptException: [string "main"]:33: attempt to get length of global 'ButtonNames' (a nil value)
The NEATevolve.lua script has a hardcoded (and relative) file reference to DP1.state. You need to make sure these files are in the same directory.
Create a Save State in BizHawk at the start of the level you want the algorithm to learn.
you’ll need to rename that file to DP1.state, and drop it in the same directory as the neatevolve.lua script. Putting both these files in the same directory as EmuHawk.exe is recommended
‘neurons’ error
LuaInterface.LuaScriptException: [string "main"]:337: attempt to index field 'neurons' (a nil value)
A similar error - try the solution above, and failing that;
As above create a quicksave at the start of a level Renamed the QuickSave1.state found in /SNES/State/ to DP1.state and move it to the folder with the EmuHawk executable.
Put the neatevolve.lua file in the same folder as EmuHawk.exe.
Noticed while I was testing that it generated a temp.pool file that seemed to have all the variables in it. Renamed that file to DP1.state.pool
‘Parameter name: source’ error
"System.ArgumentNullException: Value cannot be null. Parameter name: source"
Are you running MarI/O in a VM? Check out my notes on running MarI/O on OSX
Resources
Check out these discussions for more info on MarI/O
========================================
游戏的ROMS文件下载地址:
https://wowroms.com/en/roms/super-nintendo/super-mario-world-usa/29592.html
neatevolve.lua 文件内容:
-- MarI/O by SethBling
-- Feel free to use this code, but please do not redistribute it.
-- Intended for use with the BizHawk emulator and Super Mario World or Super Mario Bros. ROM.
-- For SMW, make sure you have a save state named "DP1.state" at the beginning of a level,
-- and put a copy in both the Lua folder and the root directory of BizHawk. if gameinfo.getromname() == "Super Mario World (USA)" then
Filename = "DP1.state"
ButtonNames = {
"A",
"B",
"X",
"Y",
"Up",
"Down",
"Left",
"Right",
}
elseif gameinfo.getromname() == "Super Mario Bros." then
Filename = "SMB1-1.state"
ButtonNames = {
"A",
"B",
"Up",
"Down",
"Left",
"Right",
}
end BoxRadius = 6
InputSize = (BoxRadius*2+1)*(BoxRadius*2+1) Inputs = InputSize+1
Outputs = #ButtonNames Population = 300
DeltaDisjoint = 2.0
DeltaWeights = 0.4
DeltaThreshold = 1.0 StaleSpecies = 15 MutateConnectionsChance = 0.25
PerturbChance = 0.90
CrossoverChance = 0.75
LinkMutationChance = 2.0
NodeMutationChance = 0.50
BiasMutationChance = 0.40
StepSize = 0.1
DisableMutationChance = 0.4
EnableMutationChance = 0.2 TimeoutConstant = 20 MaxNodes = 1000000 function getPositions()
if gameinfo.getromname() == "Super Mario World (USA)" then
marioX = memory.read_s16_le(0x94)
marioY = memory.read_s16_le(0x96) local layer1x = memory.read_s16_le(0x1A);
local layer1y = memory.read_s16_le(0x1C); screenX = marioX-layer1x
screenY = marioY-layer1y
elseif gameinfo.getromname() == "Super Mario Bros." then
marioX = memory.readbyte(0x6D) * 0x100 + memory.readbyte(0x86)
marioY = memory.readbyte(0x03B8)+16 screenX = memory.readbyte(0x03AD)
screenY = memory.readbyte(0x03B8)
end
end function getTile(dx, dy)
if gameinfo.getromname() == "Super Mario World (USA)" then
x = math.floor((marioX+dx+8)/16)
y = math.floor((marioY+dy)/16) return memory.readbyte(0x1C800 + math.floor(x/0x10)*0x1B0 + y*0x10 + x%0x10)
elseif gameinfo.getromname() == "Super Mario Bros." then
local x = marioX + dx + 8
local y = marioY + dy - 16
local page = math.floor(x/256)%2 local subx = math.floor((x%256)/16)
local suby = math.floor((y - 32)/16)
local addr = 0x500 + page*13*16+suby*16+subx if suby >= 13 or suby < 0 then
return 0
end if memory.readbyte(addr) ~= 0 then
return 1
else
return 0
end
end
end function getSprites()
if gameinfo.getromname() == "Super Mario World (USA)" then
local sprites = {}
for slot=0,11 do
local status = memory.readbyte(0x14C8+slot)
if status ~= 0 then
spritex = memory.readbyte(0xE4+slot) + memory.readbyte(0x14E0+slot)*256
spritey = memory.readbyte(0xD8+slot) + memory.readbyte(0x14D4+slot)*256
sprites[#sprites+1] = {["x"]=spritex, ["y"]=spritey}
end
end return sprites
elseif gameinfo.getromname() == "Super Mario Bros." then
local sprites = {}
for slot=0,4 do
local enemy = memory.readbyte(0xF+slot)
if enemy ~= 0 then
local ex = memory.readbyte(0x6E + slot)*0x100 + memory.readbyte(0x87+slot)
local ey = memory.readbyte(0xCF + slot)+24
sprites[#sprites+1] = {["x"]=ex,["y"]=ey}
end
end return sprites
end
end function getExtendedSprites()
if gameinfo.getromname() == "Super Mario World (USA)" then
local extended = {}
for slot=0,11 do
local number = memory.readbyte(0x170B+slot)
if number ~= 0 then
spritex = memory.readbyte(0x171F+slot) + memory.readbyte(0x1733+slot)*256
spritey = memory.readbyte(0x1715+slot) + memory.readbyte(0x1729+slot)*256
extended[#extended+1] = {["x"]=spritex, ["y"]=spritey}
end
end return extended
elseif gameinfo.getromname() == "Super Mario Bros." then
return {}
end
end function getInputs()
getPositions() sprites = getSprites()
extended = getExtendedSprites() local inputs = {} for dy=-BoxRadius*16,BoxRadius*16,16 do
for dx=-BoxRadius*16,BoxRadius*16,16 do
inputs[#inputs+1] = 0 tile = getTile(dx, dy)
if tile == 1 and marioY+dy < 0x1B0 then
inputs[#inputs] = 1
end for i = 1,#sprites do
distx = math.abs(sprites[i]["x"] - (marioX+dx))
disty = math.abs(sprites[i]["y"] - (marioY+dy))
if distx <= 8 and disty <= 8 then
inputs[#inputs] = -1
end
end for i = 1,#extended do
distx = math.abs(extended[i]["x"] - (marioX+dx))
disty = math.abs(extended[i]["y"] - (marioY+dy))
if distx < 8 and disty < 8 then
inputs[#inputs] = -1
end
end
end
end --mariovx = memory.read_s8(0x7B)
--mariovy = memory.read_s8(0x7D) return inputs
end function sigmoid(x)
return 2/(1+math.exp(-4.9*x))-1
end function newInnovation()
pool.innovation = pool.innovation + 1
return pool.innovation
end function newPool()
local pool = {}
pool.species = {}
pool.generation = 0
pool.innovation = Outputs
pool.currentSpecies = 1
pool.currentGenome = 1
pool.currentFrame = 0
pool.maxFitness = 0 return pool
end function newSpecies()
local species = {}
species.topFitness = 0
species.staleness = 0
species.genomes = {}
species.averageFitness = 0 return species
end function newGenome()
local genome = {}
genome.genes = {}
genome.fitness = 0
genome.adjustedFitness = 0
genome.network = {}
genome.maxneuron = 0
genome.globalRank = 0
genome.mutationRates = {}
genome.mutationRates["connections"] = MutateConnectionsChance
genome.mutationRates["link"] = LinkMutationChance
genome.mutationRates["bias"] = BiasMutationChance
genome.mutationRates["node"] = NodeMutationChance
genome.mutationRates["enable"] = EnableMutationChance
genome.mutationRates["disable"] = DisableMutationChance
genome.mutationRates["step"] = StepSize return genome
end function copyGenome(genome)
local genome2 = newGenome()
for g=1,#genome.genes do
table.insert(genome2.genes, copyGene(genome.genes[g]))
end
genome2.maxneuron = genome.maxneuron
genome2.mutationRates["connections"] = genome.mutationRates["connections"]
genome2.mutationRates["link"] = genome.mutationRates["link"]
genome2.mutationRates["bias"] = genome.mutationRates["bias"]
genome2.mutationRates["node"] = genome.mutationRates["node"]
genome2.mutationRates["enable"] = genome.mutationRates["enable"]
genome2.mutationRates["disable"] = genome.mutationRates["disable"] return genome2
end function basicGenome()
local genome = newGenome()
local innovation = 1 genome.maxneuron = Inputs
mutate(genome) return genome
end function newGene()
local gene = {}
gene.into = 0
gene.out = 0
gene.weight = 0.0
gene.enabled = true
gene.innovation = 0 return gene
end function copyGene(gene)
local gene2 = newGene()
gene2.into = gene.into
gene2.out = gene.out
gene2.weight = gene.weight
gene2.enabled = gene.enabled
gene2.innovation = gene.innovation return gene2
end function newNeuron()
local neuron = {}
neuron.incoming = {}
neuron.value = 0.0 return neuron
end function generateNetwork(genome)
local network = {}
network.neurons = {} for i=1,Inputs do
network.neurons[i] = newNeuron()
end for o=1,Outputs do
network.neurons[MaxNodes+o] = newNeuron()
end table.sort(genome.genes, function (a,b)
return (a.out < b.out)
end)
for i=1,#genome.genes do
local gene = genome.genes[i]
if gene.enabled then
if network.neurons[gene.out] == nil then
network.neurons[gene.out] = newNeuron()
end
local neuron = network.neurons[gene.out]
table.insert(neuron.incoming, gene)
if network.neurons[gene.into] == nil then
network.neurons[gene.into] = newNeuron()
end
end
end genome.network = network
end function evaluateNetwork(network, inputs)
table.insert(inputs, 1)
if #inputs ~= Inputs then
console.writeline("Incorrect number of neural network inputs.")
return {}
end for i=1,Inputs do
network.neurons[i].value = inputs[i]
end for _,neuron in pairs(network.neurons) do
local sum = 0
for j = 1,#neuron.incoming do
local incoming = neuron.incoming[j]
local other = network.neurons[incoming.into]
sum = sum + incoming.weight * other.value
end if #neuron.incoming > 0 then
neuron.value = sigmoid(sum)
end
end local outputs = {}
for o=1,Outputs do
local button = "P1 " .. ButtonNames[o]
if network.neurons[MaxNodes+o].value > 0 then
outputs[button] = true
else
outputs[button] = false
end
end return outputs
end function crossover(g1, g2)
-- Make sure g1 is the higher fitness genome
if g2.fitness > g1.fitness then
tempg = g1
g1 = g2
g2 = tempg
end local child = newGenome() local innovations2 = {}
for i=1,#g2.genes do
local gene = g2.genes[i]
innovations2[gene.innovation] = gene
end for i=1,#g1.genes do
local gene1 = g1.genes[i]
local gene2 = innovations2[gene1.innovation]
if gene2 ~= nil and math.random(2) == 1 and gene2.enabled then
table.insert(child.genes, copyGene(gene2))
else
table.insert(child.genes, copyGene(gene1))
end
end child.maxneuron = math.max(g1.maxneuron,g2.maxneuron) for mutation,rate in pairs(g1.mutationRates) do
child.mutationRates[mutation] = rate
end return child
end function randomNeuron(genes, nonInput)
local neurons = {}
if not nonInput then
for i=1,Inputs do
neurons[i] = true
end
end
for o=1,Outputs do
neurons[MaxNodes+o] = true
end
for i=1,#genes do
if (not nonInput) or genes[i].into > Inputs then
neurons[genes[i].into] = true
end
if (not nonInput) or genes[i].out > Inputs then
neurons[genes[i].out] = true
end
end local count = 0
for _,_ in pairs(neurons) do
count = count + 1
end
local n = math.random(1, count) for k,v in pairs(neurons) do
n = n-1
if n == 0 then
return k
end
end return 0
end function containsLink(genes, link)
for i=1,#genes do
local gene = genes[i]
if gene.into == link.into and gene.out == link.out then
return true
end
end
end function pointMutate(genome)
local step = genome.mutationRates["step"] for i=1,#genome.genes do
local gene = genome.genes[i]
if math.random() < PerturbChance then
gene.weight = gene.weight + math.random() * step*2 - step
else
gene.weight = math.random()*4-2
end
end
end function linkMutate(genome, forceBias)
local neuron1 = randomNeuron(genome.genes, false)
local neuron2 = randomNeuron(genome.genes, true) local newLink = newGene()
if neuron1 <= Inputs and neuron2 <= Inputs then
--Both input nodes
return
end
if neuron2 <= Inputs then
-- Swap output and input
local temp = neuron1
neuron1 = neuron2
neuron2 = temp
end newLink.into = neuron1
newLink.out = neuron2
if forceBias then
newLink.into = Inputs
end if containsLink(genome.genes, newLink) then
return
end
newLink.innovation = newInnovation()
newLink.weight = math.random()*4-2 table.insert(genome.genes, newLink)
end function nodeMutate(genome)
if #genome.genes == 0 then
return
end genome.maxneuron = genome.maxneuron + 1 local gene = genome.genes[math.random(1,#genome.genes)]
if not gene.enabled then
return
end
gene.enabled = false local gene1 = copyGene(gene)
gene1.out = genome.maxneuron
gene1.weight = 1.0
gene1.innovation = newInnovation()
gene1.enabled = true
table.insert(genome.genes, gene1) local gene2 = copyGene(gene)
gene2.into = genome.maxneuron
gene2.innovation = newInnovation()
gene2.enabled = true
table.insert(genome.genes, gene2)
end function enableDisableMutate(genome, enable)
local candidates = {}
for _,gene in pairs(genome.genes) do
if gene.enabled == not enable then
table.insert(candidates, gene)
end
end if #candidates == 0 then
return
end local gene = candidates[math.random(1,#candidates)]
gene.enabled = not gene.enabled
end function mutate(genome)
for mutation,rate in pairs(genome.mutationRates) do
if math.random(1,2) == 1 then
genome.mutationRates[mutation] = 0.95*rate
else
genome.mutationRates[mutation] = 1.05263*rate
end
end if math.random() < genome.mutationRates["connections"] then
pointMutate(genome)
end local p = genome.mutationRates["link"]
while p > 0 do
if math.random() < p then
linkMutate(genome, false)
end
p = p - 1
end p = genome.mutationRates["bias"]
while p > 0 do
if math.random() < p then
linkMutate(genome, true)
end
p = p - 1
end p = genome.mutationRates["node"]
while p > 0 do
if math.random() < p then
nodeMutate(genome)
end
p = p - 1
end p = genome.mutationRates["enable"]
while p > 0 do
if math.random() < p then
enableDisableMutate(genome, true)
end
p = p - 1
end p = genome.mutationRates["disable"]
while p > 0 do
if math.random() < p then
enableDisableMutate(genome, false)
end
p = p - 1
end
end function disjoint(genes1, genes2)
local i1 = {}
for i = 1,#genes1 do
local gene = genes1[i]
i1[gene.innovation] = true
end local i2 = {}
for i = 1,#genes2 do
local gene = genes2[i]
i2[gene.innovation] = true
end local disjointGenes = 0
for i = 1,#genes1 do
local gene = genes1[i]
if not i2[gene.innovation] then
disjointGenes = disjointGenes+1
end
end for i = 1,#genes2 do
local gene = genes2[i]
if not i1[gene.innovation] then
disjointGenes = disjointGenes+1
end
end local n = math.max(#genes1, #genes2) return disjointGenes / n
end function weights(genes1, genes2)
local i2 = {}
for i = 1,#genes2 do
local gene = genes2[i]
i2[gene.innovation] = gene
end local sum = 0
local coincident = 0
for i = 1,#genes1 do
local gene = genes1[i]
if i2[gene.innovation] ~= nil then
local gene2 = i2[gene.innovation]
sum = sum + math.abs(gene.weight - gene2.weight)
coincident = coincident + 1
end
end return sum / coincident
end function sameSpecies(genome1, genome2)
local dd = DeltaDisjoint*disjoint(genome1.genes, genome2.genes)
local dw = DeltaWeights*weights(genome1.genes, genome2.genes)
return dd + dw < DeltaThreshold
end function rankGlobally()
local global = {}
for s = 1,#pool.species do
local species = pool.species[s]
for g = 1,#species.genomes do
table.insert(global, species.genomes[g])
end
end
table.sort(global, function (a,b)
return (a.fitness < b.fitness)
end) for g=1,#global do
global[g].globalRank = g
end
end function calculateAverageFitness(species)
local total = 0 for g=1,#species.genomes do
local genome = species.genomes[g]
total = total + genome.globalRank
end species.averageFitness = total / #species.genomes
end function totalAverageFitness()
local total = 0
for s = 1,#pool.species do
local species = pool.species[s]
total = total + species.averageFitness
end return total
end function cullSpecies(cutToOne)
for s = 1,#pool.species do
local species = pool.species[s] table.sort(species.genomes, function (a,b)
return (a.fitness > b.fitness)
end) local remaining = math.ceil(#species.genomes/2)
if cutToOne then
remaining = 1
end
while #species.genomes > remaining do
table.remove(species.genomes)
end
end
end function breedChild(species)
local child = {}
if math.random() < CrossoverChance then
g1 = species.genomes[math.random(1, #species.genomes)]
g2 = species.genomes[math.random(1, #species.genomes)]
child = crossover(g1, g2)
else
g = species.genomes[math.random(1, #species.genomes)]
child = copyGenome(g)
end mutate(child) return child
end function removeStaleSpecies()
local survived = {} for s = 1,#pool.species do
local species = pool.species[s] table.sort(species.genomes, function (a,b)
return (a.fitness > b.fitness)
end) if species.genomes[1].fitness > species.topFitness then
species.topFitness = species.genomes[1].fitness
species.staleness = 0
else
species.staleness = species.staleness + 1
end
if species.staleness < StaleSpecies or species.topFitness >= pool.maxFitness then
table.insert(survived, species)
end
end pool.species = survived
end function removeWeakSpecies()
local survived = {} local sum = totalAverageFitness()
for s = 1,#pool.species do
local species = pool.species[s]
breed = math.floor(species.averageFitness / sum * Population)
if breed >= 1 then
table.insert(survived, species)
end
end pool.species = survived
end function addToSpecies(child)
local foundSpecies = false
for s=1,#pool.species do
local species = pool.species[s]
if not foundSpecies and sameSpecies(child, species.genomes[1]) then
table.insert(species.genomes, child)
foundSpecies = true
end
end if not foundSpecies then
local childSpecies = newSpecies()
table.insert(childSpecies.genomes, child)
table.insert(pool.species, childSpecies)
end
end function newGeneration()
cullSpecies(false) -- Cull the bottom half of each species
rankGlobally()
removeStaleSpecies()
rankGlobally()
for s = 1,#pool.species do
local species = pool.species[s]
calculateAverageFitness(species)
end
removeWeakSpecies()
local sum = totalAverageFitness()
local children = {}
for s = 1,#pool.species do
local species = pool.species[s]
breed = math.floor(species.averageFitness / sum * Population) - 1
for i=1,breed do
table.insert(children, breedChild(species))
end
end
cullSpecies(true) -- Cull all but the top member of each species
while #children + #pool.species < Population do
local species = pool.species[math.random(1, #pool.species)]
table.insert(children, breedChild(species))
end
for c=1,#children do
local child = children[c]
addToSpecies(child)
end pool.generation = pool.generation + 1 writeFile("backup." .. pool.generation .. "." .. forms.gettext(saveLoadFile))
end function initializePool()
pool = newPool() for i=1,Population do
basic = basicGenome()
addToSpecies(basic)
end initializeRun()
end function clearJoypad()
controller = {}
for b = 1,#ButtonNames do
controller["P1 " .. ButtonNames[b]] = false
end
joypad.set(controller)
end function initializeRun()
savestate.load(Filename);
rightmost = 0
pool.currentFrame = 0
timeout = TimeoutConstant
clearJoypad() local species = pool.species[pool.currentSpecies]
local genome = species.genomes[pool.currentGenome]
generateNetwork(genome)
evaluateCurrent()
end function evaluateCurrent()
local species = pool.species[pool.currentSpecies]
local genome = species.genomes[pool.currentGenome] inputs = getInputs()
controller = evaluateNetwork(genome.network, inputs) if controller["P1 Left"] and controller["P1 Right"] then
controller["P1 Left"] = false
controller["P1 Right"] = false
end
if controller["P1 Up"] and controller["P1 Down"] then
controller["P1 Up"] = false
controller["P1 Down"] = false
end joypad.set(controller)
end if pool == nil then
initializePool()
end function nextGenome()
pool.currentGenome = pool.currentGenome + 1
if pool.currentGenome > #pool.species[pool.currentSpecies].genomes then
pool.currentGenome = 1
pool.currentSpecies = pool.currentSpecies+1
if pool.currentSpecies > #pool.species then
newGeneration()
pool.currentSpecies = 1
end
end
end function fitnessAlreadyMeasured()
local species = pool.species[pool.currentSpecies]
local genome = species.genomes[pool.currentGenome] return genome.fitness ~= 0
end function displayGenome(genome)
local network = genome.network
local cells = {}
local i = 1
local cell = {}
for dy=-BoxRadius,BoxRadius do
for dx=-BoxRadius,BoxRadius do
cell = {}
cell.x = 50+5*dx
cell.y = 70+5*dy
cell.value = network.neurons[i].value
cells[i] = cell
i = i + 1
end
end
local biasCell = {}
biasCell.x = 80
biasCell.y = 110
biasCell.value = network.neurons[Inputs].value
cells[Inputs] = biasCell for o = 1,Outputs do
cell = {}
cell.x = 220
cell.y = 30 + 8 * o
cell.value = network.neurons[MaxNodes + o].value
cells[MaxNodes+o] = cell
local color
if cell.value > 0 then
color = 0xFF0000FF
else
color = 0xFF000000
end
gui.drawText(223, 24+8*o, ButtonNames[o], color, 9)
end for n,neuron in pairs(network.neurons) do
cell = {}
if n > Inputs and n <= MaxNodes then
cell.x = 140
cell.y = 40
cell.value = neuron.value
cells[n] = cell
end
end for n=1,4 do
for _,gene in pairs(genome.genes) do
if gene.enabled then
local c1 = cells[gene.into]
local c2 = cells[gene.out]
if gene.into > Inputs and gene.into <= MaxNodes then
c1.x = 0.75*c1.x + 0.25*c2.x
if c1.x >= c2.x then
c1.x = c1.x - 40
end
if c1.x < 90 then
c1.x = 90
end if c1.x > 220 then
c1.x = 220
end
c1.y = 0.75*c1.y + 0.25*c2.y end
if gene.out > Inputs and gene.out <= MaxNodes then
c2.x = 0.25*c1.x + 0.75*c2.x
if c1.x >= c2.x then
c2.x = c2.x + 40
end
if c2.x < 90 then
c2.x = 90
end
if c2.x > 220 then
c2.x = 220
end
c2.y = 0.25*c1.y + 0.75*c2.y
end
end
end
end gui.drawBox(50-BoxRadius*5-3,70-BoxRadius*5-3,50+BoxRadius*5+2,70+BoxRadius*5+2,0xFF000000, 0x80808080)
for n,cell in pairs(cells) do
if n > Inputs or cell.value ~= 0 then
local color = math.floor((cell.value+1)/2*256)
if color > 255 then color = 255 end
if color < 0 then color = 0 end
local opacity = 0xFF000000
if cell.value == 0 then
opacity = 0x50000000
end
color = opacity + color*0x10000 + color*0x100 + color
gui.drawBox(cell.x-2,cell.y-2,cell.x+2,cell.y+2,opacity,color)
end
end
for _,gene in pairs(genome.genes) do
if gene.enabled then
local c1 = cells[gene.into]
local c2 = cells[gene.out]
local opacity = 0xA0000000
if c1.value == 0 then
opacity = 0x20000000
end local color = 0x80-math.floor(math.abs(sigmoid(gene.weight))*0x80)
if gene.weight > 0 then
color = opacity + 0x8000 + 0x10000*color
else
color = opacity + 0x800000 + 0x100*color
end
gui.drawLine(c1.x+1, c1.y, c2.x-3, c2.y, color)
end
end gui.drawBox(49,71,51,78,0x00000000,0x80FF0000) if forms.ischecked(showMutationRates) then
local pos = 100
for mutation,rate in pairs(genome.mutationRates) do
gui.drawText(100, pos, mutation .. ": " .. rate, 0xFF000000, 10)
pos = pos + 8
end
end
end function writeFile(filename)
local file = io.open(filename, "w")
file:write(pool.generation .. "\n")
file:write(pool.maxFitness .. "\n")
file:write(#pool.species .. "\n")
for n,species in pairs(pool.species) do
file:write(species.topFitness .. "\n")
file:write(species.staleness .. "\n")
file:write(#species.genomes .. "\n")
for m,genome in pairs(species.genomes) do
file:write(genome.fitness .. "\n")
file:write(genome.maxneuron .. "\n")
for mutation,rate in pairs(genome.mutationRates) do
file:write(mutation .. "\n")
file:write(rate .. "\n")
end
file:write("done\n") file:write(#genome.genes .. "\n")
for l,gene in pairs(genome.genes) do
file:write(gene.into .. " ")
file:write(gene.out .. " ")
file:write(gene.weight .. " ")
file:write(gene.innovation .. " ")
if(gene.enabled) then
file:write("1\n")
else
file:write("0\n")
end
end
end
end
file:close()
end function savePool()
local filename = forms.gettext(saveLoadFile)
writeFile(filename)
end function loadFile(filename)
local file = io.open(filename, "r")
pool = newPool()
pool.generation = file:read("*number")
pool.maxFitness = file:read("*number")
forms.settext(maxFitnessLabel, "Max Fitness: " .. math.floor(pool.maxFitness))
local numSpecies = file:read("*number")
for s=1,numSpecies do
local species = newSpecies()
table.insert(pool.species, species)
species.topFitness = file:read("*number")
species.staleness = file:read("*number")
local numGenomes = file:read("*number")
for g=1,numGenomes do
local genome = newGenome()
table.insert(species.genomes, genome)
genome.fitness = file:read("*number")
genome.maxneuron = file:read("*number")
local line = file:read("*line")
while line ~= "done" do
genome.mutationRates[line] = file:read("*number")
line = file:read("*line")
end
local numGenes = file:read("*number")
for n=1,numGenes do
local gene = newGene()
table.insert(genome.genes, gene)
local enabled
gene.into, gene.out, gene.weight, gene.innovation, enabled = file:read("*number", "*number", "*number", "*number", "*number")
if enabled == 0 then
gene.enabled = false
else
gene.enabled = true
end end
end
end
file:close() while fitnessAlreadyMeasured() do
nextGenome()
end
initializeRun()
pool.currentFrame = pool.currentFrame + 1
end function loadPool()
local filename = forms.gettext(saveLoadFile)
loadFile(filename)
end function playTop()
local maxfitness = 0
local maxs, maxg
for s,species in pairs(pool.species) do
for g,genome in pairs(species.genomes) do
if genome.fitness > maxfitness then
maxfitness = genome.fitness
maxs = s
maxg = g
end
end
end pool.currentSpecies = maxs
pool.currentGenome = maxg
pool.maxFitness = maxfitness
forms.settext(maxFitnessLabel, "Max Fitness: " .. math.floor(pool.maxFitness))
initializeRun()
pool.currentFrame = pool.currentFrame + 1
return
end function onExit()
forms.destroy(form)
end writeFile("temp.pool") event.onexit(onExit) form = forms.newform(200, 260, "Fitness")
maxFitnessLabel = forms.label(form, "Max Fitness: " .. math.floor(pool.maxFitness), 5, 8)
showNetwork = forms.checkbox(form, "Show Map", 5, 30)
showMutationRates = forms.checkbox(form, "Show M-Rates", 5, 52)
restartButton = forms.button(form, "Restart", initializePool, 5, 77)
saveButton = forms.button(form, "Save", savePool, 5, 102)
loadButton = forms.button(form, "Load", loadPool, 80, 102)
saveLoadFile = forms.textbox(form, Filename .. ".pool", 170, 25, nil, 5, 148)
saveLoadLabel = forms.label(form, "Save/Load:", 5, 129)
playTopButton = forms.button(form, "Play Top", playTop, 5, 170)
hideBanner = forms.checkbox(form, "Hide Banner", 5, 190) while true do
local backgroundColor = 0xD0FFFFFF
if not forms.ischecked(hideBanner) then
gui.drawBox(0, 0, 300, 26, backgroundColor, backgroundColor)
end local species = pool.species[pool.currentSpecies]
local genome = species.genomes[pool.currentGenome] if forms.ischecked(showNetwork) then
displayGenome(genome)
end if pool.currentFrame%5 == 0 then
evaluateCurrent()
end joypad.set(controller) getPositions()
if marioX > rightmost then
rightmost = marioX
timeout = TimeoutConstant
end timeout = timeout - 1 local timeoutBonus = pool.currentFrame / 4
if timeout + timeoutBonus <= 0 then
local fitness = rightmost - pool.currentFrame / 2
if gameinfo.getromname() == "Super Mario World (USA)" and rightmost > 4816 then
fitness = fitness + 1000
end
if gameinfo.getromname() == "Super Mario Bros." and rightmost > 3186 then
fitness = fitness + 1000
end
if fitness == 0 then
fitness = -1
end
genome.fitness = fitness if fitness > pool.maxFitness then
pool.maxFitness = fitness
forms.settext(maxFitnessLabel, "Max Fitness: " .. math.floor(pool.maxFitness))
writeFile("backup." .. pool.generation .. "." .. forms.gettext(saveLoadFile))
end console.writeline("Gen " .. pool.generation .. " species " .. pool.currentSpecies .. " genome " .. pool.currentGenome .. " fitness: " .. fitness)
pool.currentSpecies = 1
pool.currentGenome = 1
while fitnessAlreadyMeasured() do
nextGenome()
end
initializeRun()
end local measured = 0
local total = 0
for _,species in pairs(pool.species) do
for _,genome in pairs(species.genomes) do
total = total + 1
if genome.fitness ~= 0 then
measured = measured + 1
end
end
end
if not forms.ischecked(hideBanner) then
gui.drawText(0, 0, "Gen " .. pool.generation .. " species " .. pool.currentSpecies .. " genome " .. pool.currentGenome .. " (" .. math.floor(measured/total*100) .. "%)", 0xFF000000, 11)
gui.drawText(0, 12, "Fitness: " .. math.floor(rightmost - (pool.currentFrame) / 2 - (timeout + timeoutBonus)*2/3), 0xFF000000, 11)
gui.drawText(100, 12, "Max Fitness: " .. math.floor(pool.maxFitness), 0xFF000000, 11)
end pool.currentFrame = pool.currentFrame + 1 emu.frameadvance();
end
==========================================================
注意:
neatevolve.lua 文件 和 DP1.State 需要放在同一目录下,不然的话执行lua脚本时会找不到游戏的起始状态文件(DP1.State)。
Super Mario World (USA).sfc 游戏文件的位置没有特殊要求,本人操作时为了方便便将其一并放在了模拟器的根目录中。
【转载】 NeuroEvolution with MarI/O —— 使用人工智能来通关超级玛丽的更多相关文章
- 马里奥AI实现方式探索 ——神经网络+增强学习
[TOC] 马里奥AI实现方式探索 --神经网络+增强学习 儿时我们都曾有过一个经典游戏的体验,就是马里奥(顶蘑菇^v^),这次里约奥运会闭幕式,日本作为2020年东京奥运会的东道主,安倍最后也已经典 ...
- [DQN] What is Deep Reinforcement Learning
已经成为DL中专门的一派,高大上的样子 Intro: MIT 6.S191 Lecture 6: Deep Reinforcement Learning Course: CS 294: Deep Re ...
- neat算法——本质就是遗传算法用于神经网络的自动构建
基于NEAT算法的马里奥AI实现 所谓NEAT算法即通过增强拓扑的进化神经网络(Evolving Neural Networks through Augmenting Topologies),算法不同 ...
- 40个超有趣的Linux命令行彩蛋和游戏
40个有趣的Linux命令行彩蛋和游戏,让你假装成日理万机的黑客高手.附一键安装脚本,在树莓派和ubuntu云主机上亲测成功,有些还可以在Windows的DOS命令行中运行. 本文配套B站视频:40个 ...
- 【转载】 Bill Gates和Elon Musk推荐,人工智能必读的三本书 -《终极算法》,《超级智能》和《终极发明》
原文地址: https://blog.csdn.net/ztf312/article/details/80761917 ---------------------------------------- ...
- 《转载》python/人工智能/Tensorflow/自然语言处理/计算机视觉/机器学习学习资源分享
本次分享一部分python/人工智能/Tensorflow/自然语言处理/计算机视觉/机器学习的学习资源,也是一些比较基础的,如果大家有看过网易云课堂的吴恩达的入门课程,在看这些视频还是一个很不错的提 ...
- 【转载】 准人工智能分享Deep Mind报告 ——AI“元强化学习”
原文地址: https://www.sohu.com/a/231895305_200424 ------------------------------------------------------ ...
- 是AI就躲个飞机-纯Python实现人工智能
你要的答案或许都在这里:小鹏的博客目录 代码下载:Here. 很久以前微信流行过一个小游戏:打飞机,这个游戏简单又无聊.在2017年来临之际,我就实现一个超级弱智的人工智能(AI),这货可以躲避从屏幕 ...
- 【翻译】用AIML实现的Python人工智能聊天机器人
前言 用python的AIML包很容易就能写一个人工智能聊天机器人. AIML是Artificial Intelligence Markup Language的简写, 但它只是一个简单的XML. 下面 ...
- 转载:[转]如何学好3D游戏引擎编程
[转]如何学好3D游戏引擎编程 Albert 本帖被 gamengines 从 游戏引擎(Game Engine) 此文为转载,但是值得一看. 此篇文章献给那些为了游戏编程不怕困难的热血青年,它的 ...
随机推荐
- Grafana 开源了一款 eBPF 采集器 Beyla
eBPF 的发展如火如荼,在可观测性领域大放异彩,Grafana 近期也发布了一款 eBPF 采集器,可以采集服务的 RED 指标,本文做一个尝鲜介绍,让读者有个大概了解. eBPF 基础介绍可以参考 ...
- ThreadLocal 源码浅析
前言 多线程在访问同一个共享变量时很可能会出现并发问题,特别是在多线程对共享变量进入写入时,那么除了加锁还有其他方法避免并发问题吗?本文将详细讲解 ThreadLocal 的使用及其源码. 一.什么是 ...
- LLM学习笔记
1. 评估榜单 1.1. C-Eval C-Eval 是一个全面的中文基础模型评估套件.它包含了13948个多项选择题,涵盖了52个不同的学科和四个难度级别. https://cevalbenchma ...
- Android日志系统(logging system)
Android日志系统(logging system) 背景 不管是做Android应用还是做Android中间层和底层,在做一些调试工作的时候,使用adb logcat非常关键.特意学习了一下安卓的 ...
- 缺氧debu模式
OxygenNotIncluded_Data(E:\SteamLibrary\steamapps\common\OxygenNotIncluded\OxygenNotIncluded_Data) 文件 ...
- NXP i.MX 8M Mini开发板规格书(四核ARM Cortex-A53 + 单核ARM Cortex-M4,主频1.6GHz)
1 评估板简介 创龙科技TLIMX8-EVM是一款基于NXP i.MX 8M Mini的四核ARM Cortex-A53 + 单核ARM Cortex-M4异构多核处理器设计的高性能评估板,由核心板和 ...
- 【Mysql】 MysqlDump导表结构或数据
mysqldump只导出表结构或只导出数据的实现方法 语法: 默认不带参数的导出,导出文本内容大概如下:创建数据库判断语句-删除表-创建表-锁表-禁用索引-插入数据-启用索引-解锁表. Usage: ...
- ubuntu pyinstaller 的安装以及使用
使用pyinstaller将Python文件打包成linux可执行文件. 安装pip3: sudo apt-get install python3-pip 更换pip3下载源: 1 sudo su 2 ...
- 蓝图中如何存储树结构: NPC对话的打开方式
BFS来扩展成数组, 然后每一个node节点的child存储为索引.
- 解锁 SQL Server 2022的时间序列数据功能
解锁 SQL Server 2022的时间序列数据功能 SQL Server2022在处理时间序列数据时,SQL Server 提供了一些优化和功能,比如 DATE_BUCKET 函数.窗口函数(如 ...
