Tree - Gradient Boosting Machine with sklearn source code
This is the second post in Boosting algorithm. In the previous post, we go through the earliest Boosting algorithm - AdaBoost, which is actually an approximation of exponential loss via additive stage-forward modelling. What if we want to choose other loss function? Can we have a more generic algorithm that can apply to all loss function.
Gradient Descent
Friedman proposed another way to optimize the additive function- Gradient descent, same as the numerical optimization method used in neural network. Basically at each step we calculate the gradient against current additive function \(F_m(x)\) to first find the direction of loss reduction and then a line search is used to find the step length.
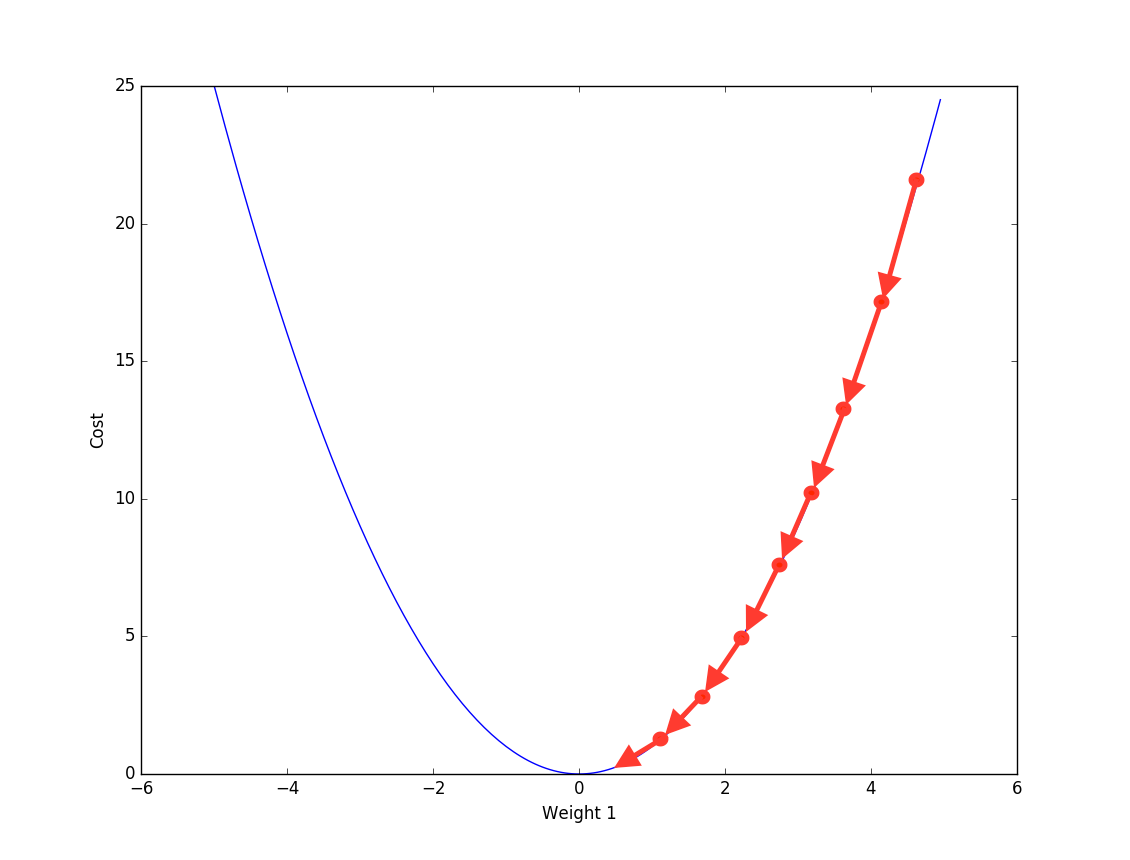
So far we have already mentioned 2 main components in Gradient Boosting machine:
- Boosting: \(F_m(x) = \sum \rho_m h(x;\alpha_m)\) final function is an additive model of multiple base learner.
- Optimization: Gradient descent is used as numeric optimization method.
There is one other important component, which we will cover later.
Gradient Boosting Basic
Here is the generic frame work of gradient Boosting algorithm.
Algorithm 1: Gradient_Boost
- \(F_0(x) = argmin \sum_i^N{L(y_i,p)}\)
- For m = 1 to M do :
A. $\hat{y_i} = - {[\frac{\partial{L(y_i, F(x_i)) }}{\partial{F(x_i ) } } ] } $ $ \text{ where \(F(x) = F_{m-1}(x)\)} $
B. \(a_m = argmin_{\alpha, \beta}\sum_i^N{[\hat{y_i} - \beta h(x_i;
\alpha)]^2}\)C. \(\rho_m = argmin_{\rho}\sum_i^N{L(y_i, F_{m-1}(x_i) + \rho h(x_i; \alpha_m))}\)
D. $ F_m(x_i) = F_{m-1}(x_i) + \rho h(x_i; \alpha_m))$
- \(F_m(x_i)\) will be final prediction
Let's go through all the above steps one by one.
Step1 we initialize the additive model, usually we can initialize with 0.
Then at each iteration:
A. Calculate negative gradient of current additive function
B. Fit a base learner to approximate negative gradient (direction of loss reduction).
C. find optimal coefficient for above base learner (step length), by minimizing the loss function.
D. update the additive function with new base learner and coefficient.
Step B is very important in the algorithm. Because given \(x\) and current additive function \(F_{m-1}(x)\), for each \(x_i\) we will have a empirical gradient. However we don't want the model to over-fit the training data. That's why an approximation estimation is used. Above we use Least-squares-Loss to fit the base leaner to the negative gradient, because least-squares estimates conditional expectation - \(E(\hat{y}|x)\)
Next let's use 2 loss function as example:
Least-Squares, all steps follow above algorithm, and we can further specify 2(A) 2(B) 2(C) as following:
Algorithm 2: LS_Boost
- \(F_0(x) = argmin \sum_i^N{L(y_i,p)}\)
- For m = 1 to M do :
A. $\hat{y_i} = y_i - F(x_i) \text{ where \(F(x) = F_{m-1}(x)\)}$
B. \(a_m = argmin_{\alpha, \beta}\sum_i^N{[\hat{y_i} - \beta h(x_i;\alpha)]^2}\)
C. $\rho_m =\beta $
D. $ F_m(x_i) = F_{m-1}(x_i) + \rho h(x_i; \alpha_m))$
Here negative gradient is simply the current residual. Since we already use least-square regression to fit base learner to the residual, where we not only get the base learner but also the coefficient. Therefore step 2(c) is no longer needed.
Least-absolute-deviation, all steps follow above algorithm, and we can further specify 2(A) 2(B) 2(C) as following:
Algorithm 3: LAD_Boost
- \(F_0(x) = argmin \sum_i^N{L(y_i,p)}\)
- For m = 1 to M do :
A. $\hat{y_i} = sign( y_i - F(x_i) ) \text{ where \(F(x) = F_{m-1}(x)\)}$
B. \(a_m = argmin_{\alpha, \beta}\sum_i^N{[\hat{y_i} - \beta h(x_i;\alpha)]^2}\)
C.\(\rho_m = argmin_{\rho}\sum_i^N{ |\hat{y_i} - \rho h(x_i; \alpha_m)|}\)
D. $ F_m(x_i) = F_{m-1}(x_i) + \rho h(x_i; \alpha_m))$
Here negative gradient is the sign of residual. And as before we fit base learner to the sign via least-squares regression.
Boosting married Tree
Now is time to reveal the last component of Gradient Boosting Machine - using Regression Tree as base learner. Same as AdaBoost, Gradient Boosting have more attractive features when it uses regression tree as base learner.
Therefore we can further represent each base leaner as an additive model (we mentioned in the previous Decision Tree Post) like below
\]
Instead of using linear regression, we fit a regression tree against negative gradient. With Least-square as loss function, \(b_j\) will the be average of gradient in each leaf.
We can further simplify this by combining \(b_j\) with coefficient \(\rho_m\), as following:
F_m(x_i) &= F_{m-1}(x_i) + \rho\sum_{j=1}^J{b_{jm} I(x \in R_{jm})}\\
F_m(x_i) &= F_{m-1}(x_i) + \sum_{j=1}^J{\lambda_{jm} I(x \in R_{jm})}
\end{align}
\]
The above transformation shows that when we fit the regression tree, we only need the node split, not the leaf assignment. In other words we are fitting Unit Gradient.
Later given the sample in each leaf, we calculate the leaf assignment by minimizing the loss function within each leaf. This will return the same result as minimizing the loss function over all sample, because all leaves are disjoint.
\]
This is also why each base learner (tree) needs to be a weak leaner. If the tree is deep enough that each leaf has only 1 sample, then we are just calculating empirical gradient of the training data (over-fitting).
From here we will use tree as default base learner, and let's go through all kinds of loss functions supported by Sklearn.
Sklean source code - loss function
Sklearn supports 7 loss function in total, 3 for classification and 4 for regression, see below
| Type | Loss | Estimator |
|---|---|---|
| ClassificationLossFunction | BinomialDeviance | LogOddsEstimator |
| ClassificationLossFunction | MultinomialDeviance | PriorProbabilityEstimator |
| ClassificationLossFunction | MultinomialDeviance | PriorProbabilityEstimator |
| RegressionLossFunction | LeastSquaresError | MeanEstimator |
| RegressionLossFunction | LeastAbsoluteError | QuantileEstimator |
| RegressionLossFunction | HuberLossFunction | QuantileEstimator |
| RegressionLossFunction | QuantileLossFunction | QuantileEstimator |
Here the Estimator calculates the prediction given training sample, including average, quantile(median), probability of each classes, or log-odds (\(\log p/1-p\)).
Each loss classes supports several methods, including calculating loss, negative gradient and update terminal regions.
For update_terminal_regions method, it first assign each sample to a leaf, perform line search to get leaf assignment and then update the prediction accordingly, see below:
class LossFunction(six.with_metaclass(ABCMeta, object)):
def update_terminal_regions(self, tree, X, y, residual, y_pred,
sample_weight, sample_mask,
learning_rate=1.0, k=0):
terminal_regions = tree.apply(X)
for leaf in np.where(tree.children_left == TREE_LEAF)[0]:
self._update_terminal_region(tree, masked_terminal_regions,
leaf, X, y, residual,
y_pred[:, k], sample_weight)
y_pred[:, k] += (learning_rate
* tree.value[:, 0, 0].take(terminal_regions, axis=0))
Next let's take a deeper dive into all loss functions.
Regression
1. Least-squares (LS)
Algorithm 4: LS_TreeBoost
- \(F_0(x) = mean(y_i)\)
- For m = 1 to M do :
A. $\hat{y_i} = y_i - F(x_i) \text{ where \(F(x) = F_{m-1}(x)\)}$
B. \(\{R_{jm}\}_1^J = \text{J terminal node tree}(\{\hat{y_i}, x_i\}_1^N)\)
C. $\lambda_{jm} = mean_{x_i \in R_{jm}}{ y_i - F(x_i)} $
D. $ F_m(x_i) = F_{m-1}(x_i) + \sum_{j=1}^J \lambda_{jm} I(x_i \in R_{jm})$
Compare algorithm 4 with algorithm 2, the difference lies in how the negative gradient is estimated, linear regression vs. regression tree. And how the coefficient is calculated, regression coefficient vs. sample mean in each leaf. And one attractive of LS is that it can get leaf assignment directly from gradient approximation. Therefore in the class update_terminal_regions is directly called to update prediction.
class LeastSquaresError(RegressionLossFunction):
def init_estimator(self):
return MeanEstimator()
def __call__(self, y, pred, sample_weight=None):
return np.mean((y - pred.ravel()) ** 2.0)
def negative_gradient(self, y, pred, **kargs):
return y - pred.ravel()
def update_terminal_regions(self, tree, X, y, residual, y_pred, learning_rate=1.0, k=0):
y_pred[:, k] += learning_rate * tree.predict(X).ravel()
2. Least-absolute-deviation (LAD)
Algorithm 5: LAD_TreeBoost
- \(F_0(x) = median(y_i)\)
- For m = 1 to M do :
A. $\hat{y_i} = sign( y_i - F(x_i) ) \text{ where \(F(x) = F_{m-1}(x)\)}$
B. \(\{R_{jm}\}_1^J = \text{J terminal node tree}(\{\hat{y_i}, x_i\}_1^N)\)
C. $\lambda_{jm} = median_{x_i \in R_{jm}}{ y_i - F(x_i)} $
D. $ F_m(x_i) = F_{m-1}(x_i) + \sum_{j=1}^J \lambda_{jm} I(x_i \in R_{jm})$
From algorithm implementation, we can see LAD is a very robust algorithm, which only use sample order to adjust the prediction accordingly. In _update_terminal_region, sample median in each leaf are used as leaf assignment.
class LeastAbsoluteError(RegressionLossFunction):
def init_estimator(self):
return QuantileEstimator(alpha=0.5)
def __call__(self, y, pred, sample_weight=None):
return np.abs(y - pred.ravel()).mean()
def negative_gradient(self, y, pred, **kargs):
pred = pred.ravel()
return 2.0 * (y - pred > 0.0) - 1.0
def _update_terminal_region(self, tree, terminal_regions, leaf, X, y, residual, pred, sample_weight):
terminal_region = np.where(terminal_regions == leaf)[0]
sample_weight = sample_weight.take(terminal_region, axis=0)
diff = y.take(terminal_region, axis=0) - pred.take(terminal_region, axis=0)
tree.value[leaf, 0, 0] = _weighted_percentile(diff, sample_weight, percentile=50)
3. Huber Loss (M-Regression)
Huber is between LS and LAD, which uses LS when the error is smaller than certain quantile and use LAD when the error exceeds certain quantile, see below:
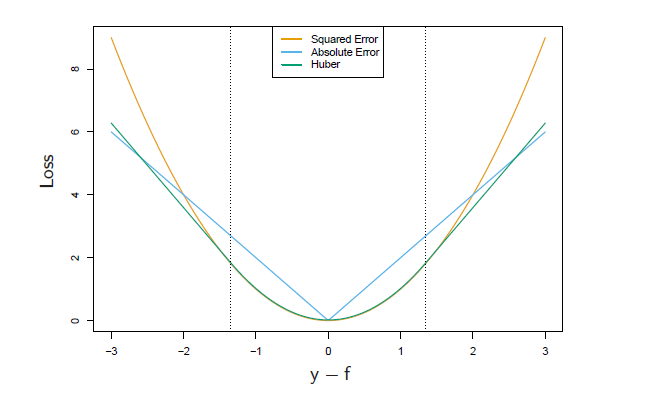
L(y,F) =
\begin{cases}
\frac{1}{2}(y-F)^2 & \quad & |y-F| \leq \delta \\
\delta(|y-F| - \delta/2) & \quad & |y-F| > \delta
\end{cases}
\end{align}
\]
where \(\delta\) is usually a certain quantile of absolute error. In additive model, we will use current additive prediction to estimate this quantile.
\]
Then we can further calculate negative gradient, and use a regression tree to approximate it.
\tilde{y} =\begin{cases}
y-F \quad & |y-F| \leq \delta \\
\delta \cdot sign(y-F) \quad & |y-F| > \delta
\end{cases}
\end{align}
\]
In the end we need to minimize loss in each leaf, which can be a little tricky with Huber loss function. For LAD, optimal leaf value is the median, for LS, optimal leaf value is the mean. For Huber an approximation method is used, where we use the sample within \(\delta\) to adjust the overall median, like following:
\]
where \(\tilde{\gamma_{jm}} = median(\gamma_{jm}) = median(y-F)\) is the LAD estimation - median of residual in each leaf.
Algorithm 6: M_TreeBoost
- \(F_0(x) = median(y_i)\)
- For m = 1 to M do :
A. \(r_{m-1} = y_i - F(x_i)\)
B. \(\delta_m = quantile_{\alpha}(|r_{m-1}|_1^N)\)
C. $
\tilde{y} =\begin{cases}
y-F \quad & |y-F| \leq \delta_m \
\delta_m \cdot sign(y-F) \quad & |y-F| > \delta_m
\end{cases} $D. \(\{R_{jm}\}_1^J = \text{J terminal node tree}(\{\tilde{y}, x_i\}_1^N)\)
E. \(\lambda_{jm}= \tilde{\gamma_{jm}} + \frac{1}{N_{jm}}\sum_{x \in R_{jm}} sign(\gamma_{m-1}(x_i) -\tilde{\gamma_{jm}} ) \cdot min(\delta_m, abs(\gamma_{m-1}(x_i) -\tilde{\gamma_{jm}}))\)
F. $ F_m(x_i) = F_{m-1}(x_i) + \sum_{j=1}^J \lambda_{jm} I(x_i \in R_{jm})$
In application, I found Huber-Loss to be a very powerful loss function. Because it is not that sensitive to the outlier, but is able to capture more information than just sample order as LAD.
class HuberLossFunction(RegressionLossFunction):
def init_estimator(self):
return QuantileEstimator(alpha=0.5)
def __call__(self, y, pred, sample_weight=None):
pred = pred.ravel()
diff = y - pred
gamma = stats.scoreatpercentile(np.abs(diff), self.alpha * 100)
gamma_mask = np.abs(diff) <= gamma
sq_loss = np.sum(0.5 * diff[gamma_mask] ** 2.0)
lin_loss = np.sum(gamma * (np.abs(diff[~gamma_mask]) - gamma / 2.0))
loss = (sq_loss + lin_loss) / y.shape[0]
return loss
def negative_gradient(self, y, pred, sample_weight=None, **kargs):
pred = pred.ravel()
diff = y - pred
gamma = stats.scoreatpercentile(np.abs(diff), self.alpha * 100)
gamma_mask = np.abs(diff) <= gamma
residual = np.zeros((y.shape[0],), dtype=np.float64)
residual[gamma_mask] = diff[gamma_mask]
residual[~gamma_mask] = gamma * np.sign(diff[~gamma_mask])
self.gamma = gamma
return residual
def _update_terminal_region(self, tree, terminal_regions, leaf, X, y,
residual, pred, sample_weight):
terminal_region = np.where(terminal_regions == leaf)[0]
sample_weight = sample_weight.take(terminal_region, axis=0)
gamma = self.gamma
diff = (y.take(terminal_region, axis=0)
- pred.take(terminal_region, axis=0))
median = _weighted_percentile(diff, sample_weight, percentile=50)
diff_minus_median = diff - median
tree.value[leaf, 0] = median + np.mean(
np.sign(diff_minus_median) *
np.minimum(np.abs(diff_minus_median), gamma))
Classification
1. 2-class classification
For binomial classification, our calculation below will be slightly different from Freud's paper. Because Sklearn use \(y \in \{0,1\}\), while in Freud's paper \(y \in \{-1,1\}\) is used. Of course, whichever you use, you should get the same result. You need to make sure that your calculation is consistent. Relevant bug was spotted in Sklearn before.
Do you still recall how is the binomial log-likelihood defined? We define current prediction as log-odds.
F & = \log(\frac{P(y=1|x)}{P(y=0|x)}) \\
P &= \frac{1}{1+e^{-F}} \\
\end{align}
\]
And use above to calculate negative log-likelihood function, we get following
L(y,F) & = - E(y\log{p} + (1-y)log{(1-p)} ) \\
& = - E( y\log{\frac{p}{1-p}} + \log({1-p}) ) \\
& = - E( yF - \log{(1+e^F)} )
\end{align}
\]
The negative gradient is below, I also give a general version, which is in line with later k-class regression
\tilde{y} & = y - \frac{1}{1+ e^{-,F}} = y - \sigma(F) = y - p
\end{align}
\]
In the end we calculate leaf assignment by minimizing the loss in each leaf
\]
There is no close solution to above function, a second-order Newton Raphson is used to approximate
Quick Note - Newton Raphson
\(f(x + \epsilon) \approx f(x) + f'(x)\epsilon + \frac{1}{2}f''(x)\epsilon^2\) 2nd order Taylor expansion
To get optimal value, \(f'(x) = 0\)
we will get \(\epsilon = -\frac{f'(x)}{f''(x)}\)
\lambda_{jm} & = \sum_{x \in R_{jm}}(y - \frac{1}{1+ e^{-F}} )/(\frac{1}{1+ e^{-F}} \cdot \frac{1}{1+ e^{F}}) \\
& = \sum_{x \in R_{jm}} \tilde{y} / ( (y-\tilde{y}) \cdot (1-y+\tilde{y}) )
\end{align}
\]
Algorithm 7: L2_TreeBoost
- \(F_0(x) = 0\)
- For m = 1 to M do :
A. $\tilde{y} = y - \frac{1}{1+ e^{-F}} \text{ where \(F(x) = F_{m-1}(x)\)}$
B. \(\{R_{jm}\}_1^J = \text{J terminal node tree}(\{\hat{y_i}, x_i\}_1^N)\)
C. $\lambda_{jm} = \sum_{x \in R_{jm}} \tilde{y} / ( (y-\tilde{y}) \cdot (1-y+\tilde{y})) $
D. $ F_m(x_i) = F_{m-1}(x_i) + \sum_{j=1}^J \lambda_{jm} I(x_i \in R_{jm})$
class BinomialDeviance(ClassificationLossFunction):
def init_estimator(self):
return LogOddsEstimator()
def __call__(self, y, pred, sample_weight=None):
pred = pred.ravel()
return -2.0 * np.mean((y * pred) - np.logaddexp(0.0, pred)) #logaddexp(0, v) == log(1.0 + exp(v))
def negative_gradient(self, y, pred, **kargs):
return y - expit(pred.ravel()) # sigmoid function
def _update_terminal_region(self, tree, terminal_regions, leaf, X, y,
residual, pred, sample_weight):
terminal_region = np.where(terminal_regions == leaf)[0]
residual = residual.take(terminal_region, axis=0)
y = y.take(terminal_region, axis=0)
sample_weight = sample_weight.take(terminal_region, axis=0)
numerator = np.sum(sample_weight * residual)
denominator = np.sum(sample_weight * (y - residual) * (1 - y + residual))
tree.value[leaf, 0, 0] = numerator / denominator
2. k-class classification
Loss function is defined in the same ways with multiclass:
\]
where \(p_k(x) = exp(F_k(x))/\sum_{l=1}^Kexp(F_l(x))\)
Therefore we will get the loss function nad negative gradient as following:
L(y,F) &= -\sum_{k=1}^K y_kF_k(x) - log(\sum_{l=1}^Kexp(F_l(x)))\\
\tilde{y_i} &= y_{ki} - p_{k,m-1}(x_i)
\end{align}
\]
Algorithm 8: LK_TreeBoost
- \(F_0(x) = 0\)
- For m = 1 to M do :
A. \(p_k(x) = exp(F_k(x))/\sum_{l=1}^Kexp(F_l(x))\)
B. \(\tilde{y_i} = y_i - p_k(x_i)\)
B. \(\{R_{jm}\}_1^J = \text{J terminal node tree}(\{\hat{y_i}, x_i\}_1^N)\)
C. $\lambda_{jm} = \sum_{x \in R_{jm}} \tilde{y} / ( (y-\tilde{y}) \cdot (1-y+\tilde{y})) $
D. $ F_m(x_i) = F_{m-1}(x_i) + \sum_{j=1}^J \lambda_{jm} I(x_i \in R_{jm})$
sklearn source code - GBM Framework
Base Learner
Base learner in each iteration is trained via fit_stage method. A Decision Tree is trained to approximate negative gradient given current additive function. And then leaf assignment is calculated to minimize loss function in each leaf.
def _fit_stage(self, i, X, y, y_pred, sample_weight, sample_mask, random_state, X_idx_sorted, X_csc=None, X_csr=None):
assert sample_mask.dtype == np.bool
loss = self.loss_
original_y = y
for k in range(loss.K):
residual = loss.negative_gradient(y, y_pred, k=k, sample_weight=sample_weight)
tree = DecisionTreeRegressor(
criterion=self.criterion,
splitter='best',
max_depth=self.max_depth,
min_samples_split=self.min_samples_split,
min_samples_leaf=self.min_samples_leaf,
min_weight_fraction_leaf=self.min_weight_fraction_leaf,
min_impurity_decrease=self.min_impurity_decrease,
min_impurity_split=self.min_impurity_split,
max_features=self.max_features,
max_leaf_nodes=self.max_leaf_nodes,
random_state=random_state,
presort=self.presort)
tree.fit(X, residual, sample_weight=sample_weight,
check_input=False, X_idx_sorted=X_idx_sorted)
loss.update_terminal_regions(tree.tree_, X, y, residual, y_pred,sample_weight, sample_mask, self.learning_rate, k=k)
self.estimators_[i, k] = tree
return y_pred
Boosting
Boosting is performed upon above base learner via fit_stages method. In each iteration a new base learner is trained given current additive function.
n_estimators specifies the number of iteration (# of base learner).
subsample indicates the split between training sample and validation sample. Base learner is trained against training sample, and we use untouched validation sample for performance.
def _fit_stages(self, X, y, y_pred, sample_weight, random_state,
begin_at_stage=0, monitor=None, X_idx_sorted=None):
n_samples = X.shape[0]
do_oob = self.subsample < 1.0
sample_mask = np.ones((n_samples, ), dtype=np.bool)
n_inbag = max(1, int(self.subsample * n_samples))
loss_ = self.loss_
i = begin_at_stage
for i in range(begin_at_stage, self.n_estimators):
if do_oob:
sample_mask = _random_sample_mask(n_samples, n_inbag,
random_state)
old_oob_score = loss_(y[~sample_mask],
y_pred[~sample_mask],
sample_weight[~sample_mask])
y_pred = self._fit_stage(i, X, y, y_pred, sample_weight, sample_mask, random_state, X_idx_sorted, X_csc, X_csr)
if do_oob:
self.train_score_[i] = loss_(y[sample_mask],
y_pred[sample_mask], sample_weight[sample_mask])
self.oob_improvement_[i] = (
old_oob_score - loss_(y[~sample_mask],
y_pred[~sample_mask], sample_weight[~sample_mask]))
return i + 1
Reference
- J. Friedman, Greedy Function Approximation: A Gradient Boosting Machine, The Annals of Statistics, Vol. 29, No. 5, 2001.
- J. Friedman, Stochastic Gradient Boosting, 1999
- T. Hastie, R. Tibshirani and J. Friedman. Elements of Statistical Learning Ed. 2, Springer, 2009.
- Bishop, Pattern Recognition and Machine Learning 2006
- scikit-learn tutorial http://scikit-learn.org/stable/modules/generated/sklearn.ensemble.GradientBoostingClassifier.html
Tree - Gradient Boosting Machine with sklearn source code的更多相关文章
- Tree - AdaBoost with sklearn source code
In the previous post we addressed some issue of decision tree, including instability, lack of smooth ...
- Tree - Decision Tree with sklearn source code
After talking about Information theory, now let's come to one of its application - Decision Tree! No ...
- 论文笔记:GREEDY FUNCTION APPROXIMATION: A GRADIENT BOOSTING MACHINE
Boost是集成学习方法中的代表思想之一,核心的思想是不断的迭代.boost通常采用改变训练数据的概率分布,针对不同的训练数据分布调用弱学习算法学习一组弱分类器.在多次迭代的过程中,当前次迭代所用的训 ...
- Python中Gradient Boosting Machine(GBM)调参方法详解
原文地址:Complete Guide to Parameter Tuning in Gradient Boosting (GBM) in Python by Aarshay Jain 原文翻译与校对 ...
- 机器学习--Gradient Boosting Machine(GBM)调参方法详解
一.GBM参数 总的来说GBM的参数可以被归为三类: 树参数:调节模型中每个决策树的性质 Boosting参数:调节模型中boosting的操作 其他模型参数:调节模型总体的各项运作 1.树参数 现在 ...
- Greedy Function Approximation:A Gradient Boosting Machine
https://statweb.stanford.edu/~jhf/ftp/trebst.pdf page10 90% to 95% of the observations were often de ...
- A Gentle Introduction to the Gradient Boosting Algorithm for Machine Learning
A Gentle Introduction to the Gradient Boosting Algorithm for Machine Learning by Jason Brownlee on S ...
- Gradient Boosting Decision Tree学习
Gradient Boosting Decision Tree,即梯度提升树,简称GBDT,也叫GBRT(Gradient Boosting Regression Tree),也称为Multiple ...
- How to Configure the Gradient Boosting Algorithm
How to Configure the Gradient Boosting Algorithm by Jason Brownlee on September 12, 2016 in XGBoost ...
随机推荐
- 获取url中?后面传递的参数
function getUrlArgs(){ var url=location.href; var i=url.indexOf('?'); if(i==-1)return; var querystr= ...
- IO流C++
1.iostream处理控制台IO #include<iostream> #include<string> using namespace std; istream& ...
- Java代码生成器CodeX4J介绍
用代码生成器生成一些固定的或有规律的代码,可以有效的提高工作效率.我写了个开源的,放在了GitHub上,希望能对有需要的朋友有所帮助. GitHub代码地址https://github.com/jac ...
- Scala 语法基础
一 简介 Scala 是一门多范式(multi-paradigm)的编程语言,设计初衷是要集成面向对象编程和函数式编程的各种特性.Scala 运行在Java虚拟机上,并兼容现有的Java程序.Scal ...
- ajaxSubmit请求返回数据成功,但是不执行success回调函数
最近项目涉及到附件上传就头痛,一直在用plupload插件在做...ie9偶尔抽风但还是可以的... 然后有个需求,表格每行都有个上传按钮,页面多上传按钮. 一.开始的时候,用plupload做的,多 ...
- Struts2速记手册
工作原理 Action类 Action类 普通Action类 私有属性及getter.setter(处理请求参数) execute()方法(处理请求) 实现Action接口 提供常量 继承Ac ...
- Redis高可用详解:持久化技术及方案选择 (推荐)--转载自编程迷思博客www.cnblogs.com/kismetv/p/8654978.html
一.Redis高可用概述 在介绍Redis高可用之前,先说明一下在Redis的语境中高可用的含义. 我们知道,在web服务器中,高可用是指服务器可以正常访问的时间,衡量的标准是在多长时间内可以提供正常 ...
- java 加载并读取Properties 文件
1 .系统自带的application.properties (以下代码仅供参考,不能粘贴复制) 假设application.properties文件有下面两个值: come.test.name = ...
- Delphi 高效读写锁
本人设计了一个高效读写锁,可实现多个线程读一个线程写的锁,应该比Delphi自带的读写锁高效,本人没有做对比测试. 本文的锁不可以在一个线程里重入,否则会锁死,另外读写锁最多支持65535个线程同时读 ...
- C++中一个0xC0000005访问冲突问题
在冯.诺依曼结构CPU(如i386,ARM A8,A9)的保护模式或者哈佛结构(如8051, ARM M0,.. M3)的CPU下,C++编译器将放置常量的内存设置为只读模式或者放入只读内存中,如果出 ...
