吴裕雄--天生自然 PYTHON数据分析:威斯康星乳腺癌(诊断)数据分析(续一)
drop_list1 = ['perimeter_mean','radius_mean','compactness_mean','concave points_mean','radius_se','perimeter_se','radius_worst','perimeter_worst','compactness_worst','concave points_worst','compactness_se','concave points_se','texture_worst','area_worst']
x_1 = x.drop(drop_list1,axis = 1 ) # do not modify x, we will use it later
x_1.head()
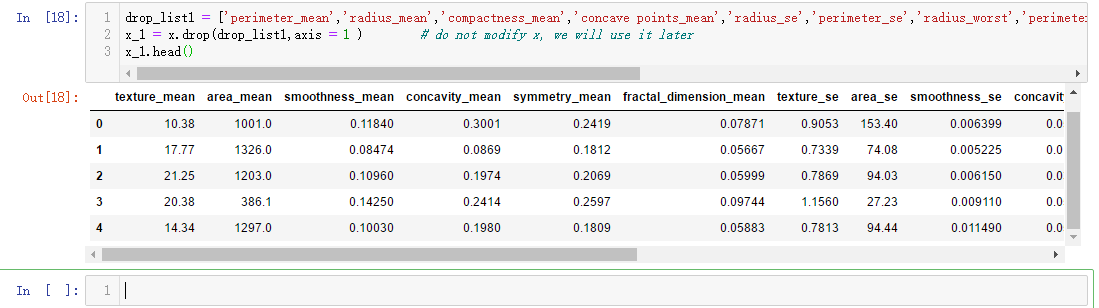
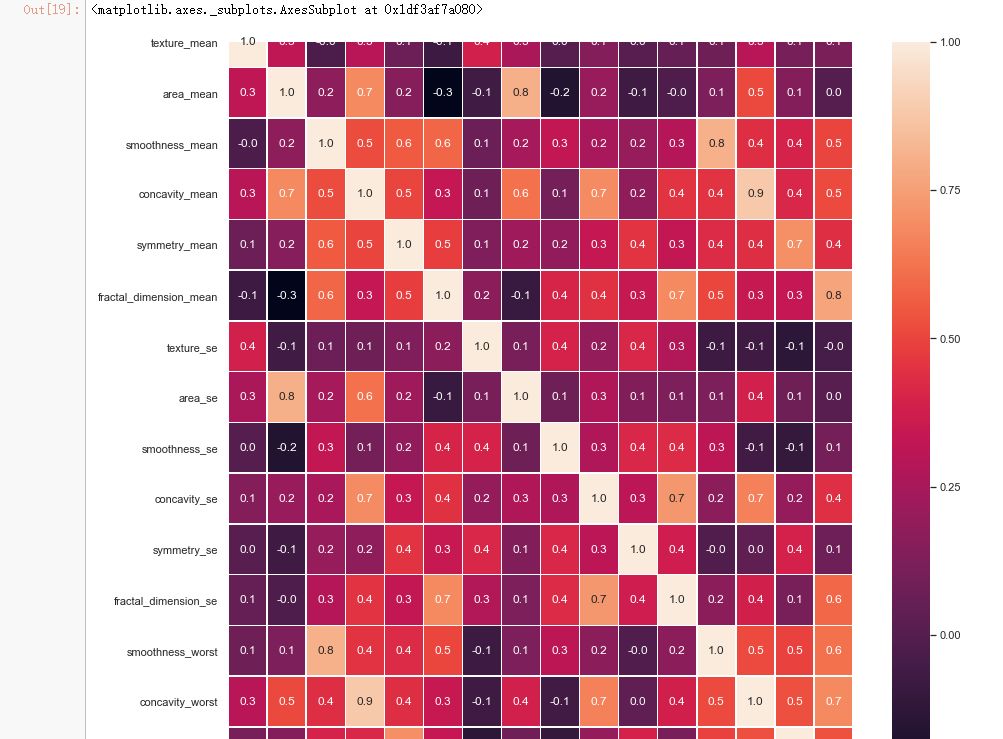
#correlation map
f,ax = plt.subplots(figsize=(14, 14))
sns.heatmap(x_1.corr(), annot=True, linewidths=.5, fmt= '.1f',ax=ax)
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import f1_score,confusion_matrix
from sklearn.metrics import accuracy_score # split data train 70 % and test 30 %
x_train, x_test, y_train, y_test = train_test_split(x_1, y, test_size=0.3, random_state=42) #random forest classifier with n_estimators=10 (default)
clf_rf = RandomForestClassifier(random_state=43)
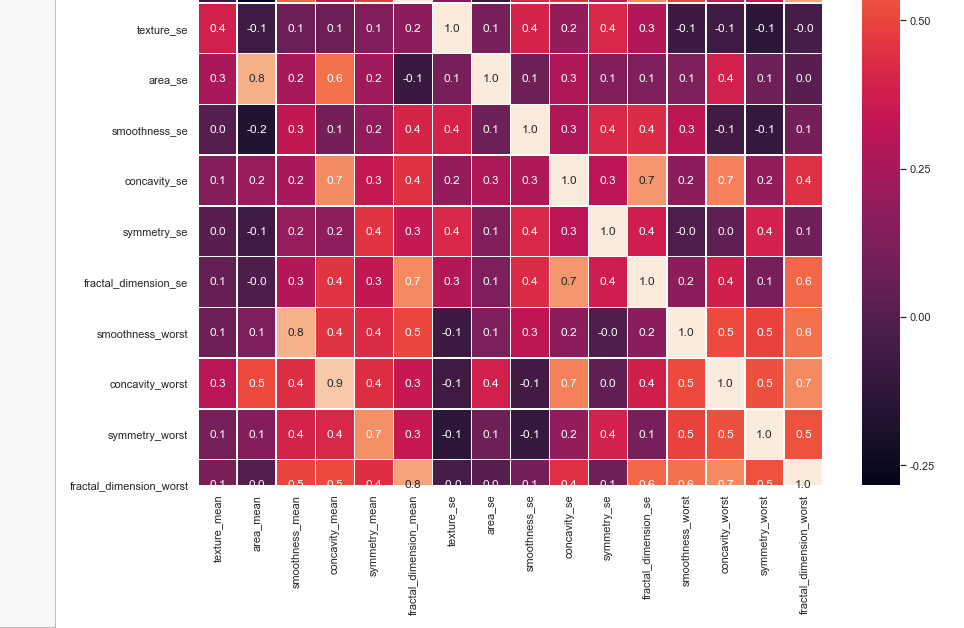
clr_rf = clf_rf.fit(x_train,y_train) ac = accuracy_score(y_test,clf_rf.predict(x_test))
print('Accuracy is: ',ac)
cm = confusion_matrix(y_test,clf_rf.predict(x_test))
sns.heatmap(cm,annot=True,fmt="d")
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
# find best scored 5 features
select_feature = SelectKBest(chi2, k=5).fit(x_train, y_train)
print('Score list:', select_feature.scores_)
print('Feature list:', x_train.columns)

x_train_2 = select_feature.transform(x_train)
x_test_2 = select_feature.transform(x_test)
#random forest classifier with n_estimators=10 (default)
clf_rf_2 = RandomForestClassifier()
clr_rf_2 = clf_rf_2.fit(x_train_2,y_train)
ac_2 = accuracy_score(y_test,clf_rf_2.predict(x_test_2))
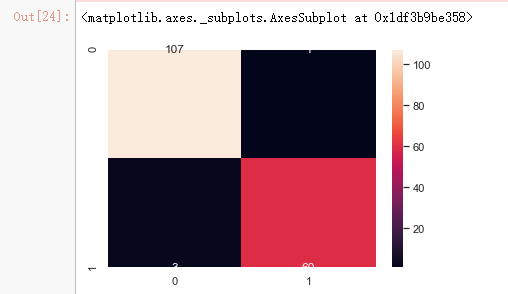
print('Accuracy is: ',ac_2)
cm_2 = confusion_matrix(y_test,clf_rf_2.predict(x_test_2))
sns.heatmap(cm_2,annot=True,fmt="d")
from sklearn.feature_selection import RFE
# Create the RFE object and rank each pixel
clf_rf_3 = RandomForestClassifier()
rfe = RFE(estimator=clf_rf_3, n_features_to_select=5, step=1)
rfe = rfe.fit(x_train, y_train)
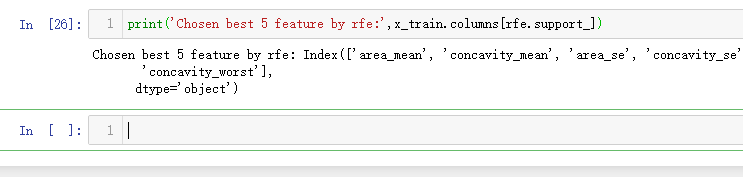
print('Chosen best 5 feature by rfe:',x_train.columns[rfe.support_])
from sklearn.feature_selection import RFECV # The "accuracy" scoring is proportional to the number of correct classifications
clf_rf_4 = RandomForestClassifier()
rfecv = RFECV(estimator=clf_rf_4, step=1, cv=5,scoring='accuracy') #5-fold cross-validation
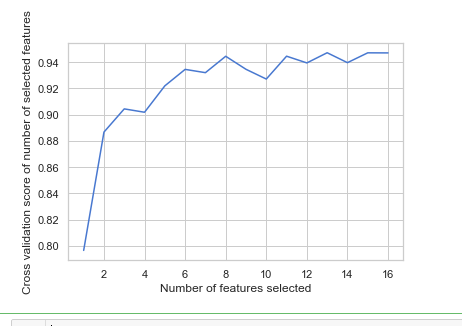
rfecv = rfecv.fit(x_train, y_train) print('Optimal number of features :', rfecv.n_features_)
print('Best features :', x_train.columns[rfecv.support_])
# Plot number of features VS. cross-validation scores
import matplotlib.pyplot as plt
plt.figure()
plt.xlabel("Number of features selected")
plt.ylabel("Cross validation score of number of selected features")
plt.plot(range(1, len(rfecv.grid_scores_) + 1), rfecv.grid_scores_)
plt.show()
clf_rf_5 = RandomForestClassifier()
clr_rf_5 = clf_rf_5.fit(x_train,y_train)
importances = clr_rf_5.feature_importances_
std = np.std([tree.feature_importances_ for tree in clf_rf.estimators_],
axis=0)
indices = np.argsort(importances)[::-1] # Print the feature ranking
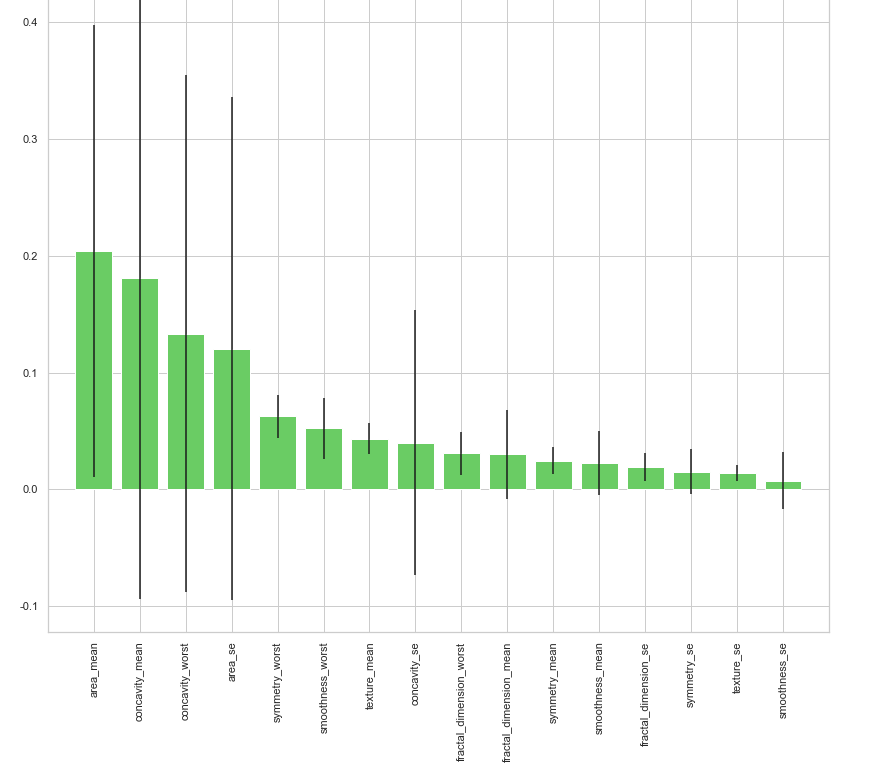
print("Feature ranking:") for f in range(x_train.shape[1]):
print("%d. feature %d (%f)" % (f + 1, indices[f], importances[indices[f]])) # Plot the feature importances of the forest plt.figure(1, figsize=(14, 13))
plt.title("Feature importances")
plt.bar(range(x_train.shape[1]), importances[indices],
color="g", yerr=std[indices], align="center")
plt.xticks(range(x_train.shape[1]), x_train.columns[indices],rotation=90)
plt.xlim([-1, x_train.shape[1]])
plt.show()

# split data train 70 % and test 30 %
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.3, random_state=42)
#normalization
x_train_N = (x_train-x_train.mean())/(x_train.max()-x_train.min())
x_test_N = (x_test-x_test.mean())/(x_test.max()-x_test.min()) from sklearn.decomposition import PCA
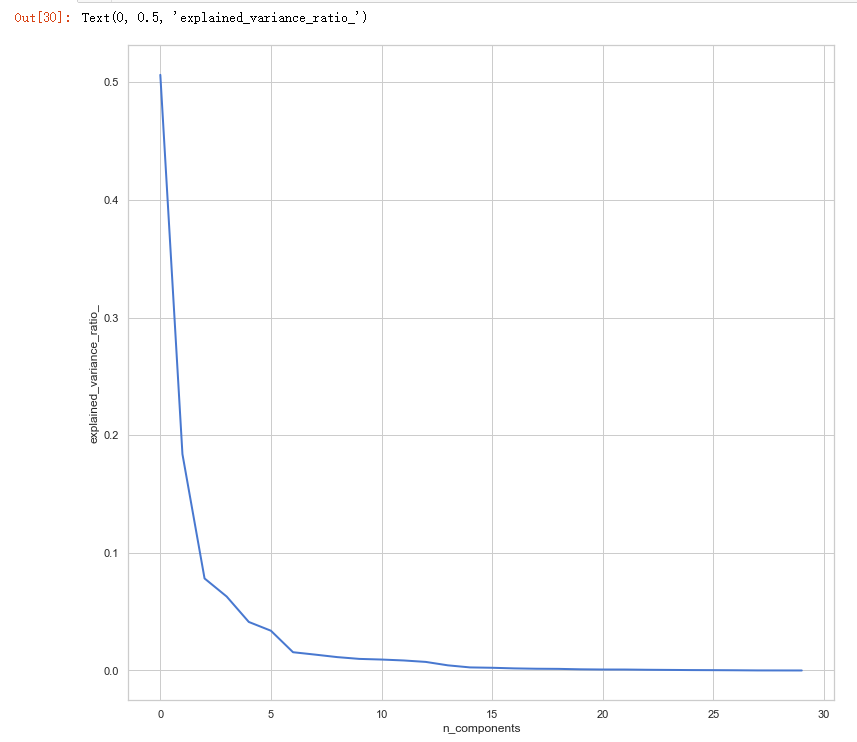
pca = PCA()
pca.fit(x_train_N) plt.figure(1, figsize=(14, 13))
plt.clf()
plt.axes([.2, .2, .7, .7])
plt.plot(pca.explained_variance_ratio_, linewidth=2)
plt.axis('tight')
plt.xlabel('n_components')
plt.ylabel('explained_variance_ratio_')
吴裕雄--天生自然 PYTHON数据分析:威斯康星乳腺癌(诊断)数据分析(续一)的更多相关文章
- 吴裕雄--天生自然 PYTHON数据分析:糖尿病视网膜病变数据分析(完整版)
# This Python 3 environment comes with many helpful analytics libraries installed # It is defined by ...
- 吴裕雄--天生自然 PYTHON数据分析:所有美国股票和etf的历史日价格和成交量分析
# This Python 3 environment comes with many helpful analytics libraries installed # It is defined by ...
- 吴裕雄--天生自然 python数据分析:健康指标聚集分析(健康分析)
# This Python 3 environment comes with many helpful analytics libraries installed # It is defined by ...
- 吴裕雄--天生自然 python数据分析:葡萄酒分析
# import pandas import pandas as pd # creating a DataFrame pd.DataFrame({'Yes': [50, 31], 'No': [101 ...
- 吴裕雄--天生自然 PYTHON数据分析:人类发展报告——HDI, GDI,健康,全球人口数据数据分析
import pandas as pd # Data analysis import numpy as np #Data analysis import seaborn as sns # Data v ...
- 吴裕雄--天生自然 python数据分析:医疗费数据分析
import numpy as np import pandas as pd import os import matplotlib.pyplot as pl import seaborn as sn ...
- 吴裕雄--天生自然 PYTHON语言数据分析:ESA的火星快车操作数据集分析
import os import numpy as np import pandas as pd from datetime import datetime import matplotlib imp ...
- 吴裕雄--天生自然 python语言数据分析:开普勒系外行星搜索结果分析
import pandas as pd pd.DataFrame({'Yes': [50, 21], 'No': [131, 2]}) pd.DataFrame({'Bob': ['I liked i ...
- 吴裕雄--天生自然 PYTHON数据分析:基于Keras的CNN分析太空深处寻找系外行星数据
#We import libraries for linear algebra, graphs, and evaluation of results import numpy as np import ...
随机推荐
- 基于node的前后端分离初识
久闻node的大名,先后也看过node的文档,但是,总是碍于没有挑起我的G点,所以实际codeing的例子都没有.最近,突然很有兴致,想把原有用页面ajax渲染的页面采用服务端node来渲染,研究了两 ...
- django的认证演变过程分析
认证规则图: django不分离 drf分类 认证规则演变图 数据库session认证:低效 缓存认证:高效 jwt认证:高效 缓存认证:不易并发 jwt认证:易并发
- IDEA中的常用插件安装以及使用的介绍
IDEA中的lombok插件安装以及各注解的详细介绍 Grep Console 当你密密麻麻一大片的日志,去查看起来,很容易看花眼:这个工具正好解决了这个痛点,可以说它就是 IDEA 自带 Conso ...
- $.post系统登录校验
AJAX方式:$.post系统登录校验 https://blog.csdn.net/woshisangsang/article/details/66560238?utm_source=blogxgwz ...
- 如何找到我的Python site-packages目录的位置?
来源:广州SEO 我如何找到我的site-packages目录的位置? #1楼 这对我有用: python -m site --user-site #2楼 从“如何安装Django”文档 (尽管这不仅 ...
- C3D使用指南
C3D GitHub项目地址:https://github.com/facebook/C3D C3D 官方用户指南:https://goo.gl/k2SnLY 1. C3D特征提取 1.1 命令参数介 ...
- java replaceall 用法:处理特殊字符
public class TryDotRegEx { public static void main(String[] args) { // TODO Auto-generated method st ...
- Office customUI中如何动态更新控件标题和图标?
本例,在Excel右键菜单中创建一个按钮,按钮的标题使用getLabel动态获取,图标使用getImage动态获取. customUI XML代码: <customUI xmlns=" ...
- numpy模块介绍
import numpy as np np.array([1,2,3]) array([1, 2, 3]) np.array([[1,2,3],[4,5,6]]) array([[1, 2, 3], ...
- 量化预测质量之分类报告 sklearn.metrics.classification_report
classification_report的调用为:classification_report(y_true, y_pred, labels=None, target_names=None, samp ...
