dp-(LCS 基因匹配)
| Time Limit: 1000MS | Memory Limit: 10000K | |
| Total Submissions: 19885 | Accepted: 11100 |
Description
A human gene can be identified through a series of
time-consuming biological experiments, often with the help of computer
programs. Once a sequence of a gene is obtained, the next job is to
determine its function.
One of the methods for biologists to use in determining the function
of a new gene sequence that they have just identified is to search a
database with the new gene as a query. The database to be searched
stores many gene sequences and their functions – many researchers have
been submitting their genes and functions to the database and the
database is freely accessible through the Internet.
A database search will return a list of gene sequences from the database that are similar to the query gene.
Biologists assume that sequence similarity often implies
functional similarity. So, the function of the new gene might be
one of the functions that the genes from the list have. To exactly
determine which one is the right one another series of biological
experiments will be needed.
Your job is to make a program that compares two genes and determines
their similarity as explained below. Your program may be used as a part
of the database search if you can provide an efficient one.
Given two genes AGTGATG and GTTAG, how similar are they? One of the methods to measure the similarity
of two genes is called alignment. In an alignment, spaces are inserted, if necessary, in appropriate positions of
the genes to make them equally long and score the resulting genes according to a scoring matrix.
For example, one space is inserted into AGTGATG to result in
AGTGAT-G, and three spaces are inserted into GTTAG to result in
–GT--TAG. A space is denoted by a minus sign (-). The two genes
are now of equal
length. These two strings are aligned:
AGTGAT-G
-GT--TAG
In this alignment, there are four matches, namely, G in the second
position, T in the third, T in the sixth, and G in the eighth. Each
pair of aligned characters is assigned a score according to the
following scoring matrix.
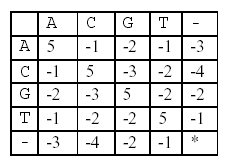
denotes that a space-space match is not allowed. The score
of the alignment above is (-3)+5+5+(-2)+(-3)+5+(-3)+5=9.
Of course, many other alignments are possible. One is shown below (a
different number of spaces are inserted into different positions):
AGTGATG
-GTTA-G
This alignment gives a score of (-3)+5+5+(-2)+5+(-1) +5=14. So,
this one is better than the previous one. As a matter of fact, this one
is optimal since no other alignment can have a higher score. So, it is
said that the
similarity of the two genes is 14.
Input
input consists of T test cases. The number of test cases ) (T is
given in the first line of the input file. Each test case consists of
two lines: each line contains an integer, the length of a gene, followed
by a gene sequence. The length of each gene sequence is at least one
and does not exceed 100.
Output
Sample Input
2
7 AGTGATG
5 GTTAG
7 AGCTATT
9 AGCTTTAAA
Sample Output
14
21 题目大意 : 给出一个基因匹配表格 , 里面有一些数值 。
设 dp[i][j] 为 s1 取第 i 个字符, s2 取第 j 个字符的最大值,决定dp[i][j] 最优的情况有 三种, 类似于最长公共子序列的三种情况。
1 . s1 取第 i 个字母, s2 取 '-' , temp1 = dp[i-1][j] + score[a[i]]['-'];
2 . s2 取第 j 个字母, s1 取 '-' , temp2 = dp[i][j-1] + score['-'][b[j]];
3 . s1 取第 i 个字母 , s2 取第 j 个字母 , temp3 = dp[i-1][j-1] + score[a[i]][b[j]]; 则 dp[i][j] = max ( temp1, temp2, temp3 ); 还有初始化问题 : dp[0][0] = 0;
dp[i][0] = dp[i-1][0] + score[a[i]]['-'];
dp[0][j] = dp[0][j-1] + score['-'][b[j]]; 代码示例 :
/*
* Author: ry
* Created Time: 2017/9/3 8:00:06
* File Name: 1.cpp
*/
#include <iostream>
#include <cstdio>
#include <cstdlib>
#include <cstring>
#include <cmath>
#include <algorithm>
#include <string>
#include <vector>
#include <stack>
#include <queue>
#include <set>
#include <time.h>
using namespace std;
const int mm = 1e6+5;
#define ll long long ll t_cnt;
void t_st(){t_cnt=clock();}
void t_ot(){printf("you spent : %lldms\n", clock()-t_cnt);}
//开始t_st();
//结束t_ot(); int dp[150][150];
int score['T'+1]['T'+1]; void intial () {
score['A']['A'] = 5;
score['C']['C'] = 5;
score['G']['G'] = 5;
score['T']['T'] = 5;
score['A']['C'] = score['C']['A'] = -1;
score['A']['G'] = score['G']['A'] = -2;
score['A']['T'] = score['T']['A'] = -1;
score['A']['-'] = score['-']['A'] = -3;
score['C']['G'] = score['G']['C'] = -3;
score['C']['T'] = score['T']['C'] = -2;
score['C']['-'] = score['-']['C'] = -4;
score['G']['T'] = score['T']['G'] = -2;
score['G']['-'] = score['-']['G'] = -2;
score['T']['-'] = score['-']['T'] = -1;
} int MAX (int x, int y, int z) {
int k = (x>y?x:y);
return z>k?z:k;
} int main() {
int t;
int len1, len2;
char a[105], b[105]; intial();
cin >> t;
getchar();
while ( t-- ){
memset (dp, 0, sizeof(dp));
scanf("%d %s", &len1, a);
scanf("%d %s", &len2, b); for (int i = len1; i > 0; i--)
a[i] = a[i-1];
for (int i = len2; i > 0; i--)
b[i] = b[i-1]; dp[0][0] = 0;
for (int i = 1; i <= len1; i++)
dp[i][0] = dp[i-1][0] + score[a[i]]['-'];
for (int j = 1; j <= len2; j++)
dp[0][j] = dp[0][j-1] + score['-'][b[j]]; for (int i = 1; i <= len1; i++)
for (int j = 1; j <= len2; j++){
int temp1 = dp[i-1][j] + score[a[i]]['-'];
int temp2 = dp[i][j-1] + score['-'][b[j]];
int temp3 = dp[i-1][j-1] + score[a[i]][b[j]];
dp[i][j] = MAX (temp1, temp2, temp3);
} printf ("%d\n", dp[len1][len2]);
} return 0;
}
dp-(LCS 基因匹配)的更多相关文章
- 【线型DP】【LCS】洛谷P4303 [AHOI2006]基因匹配
P4303 [AHOI2006]基因匹配 标签(空格分隔): 考试题 nt题 LCS优化 [题目] 卡卡昨天晚上做梦梦见他和可可来到了另外一个星球,这个星球上生物的DNA序列由无数种碱基排列而成(地球 ...
- BZOJ 1264: [AHOI2006]基因匹配Match( LCS )
序列最大长度2w * 5 = 10w, O(n²)的LCS会T.. LCS 只有当a[i] == b[j]时, 才能更新答案, 我们可以记录n个数在第一个序列中出现的5个位置, 然后从左往右扫第二个序 ...
- bzoj 1264 [AHOI2006]基因匹配Match(DP+树状数组)
1264: [AHOI2006]基因匹配Match Time Limit: 10 Sec Memory Limit: 162 MBSubmit: 793 Solved: 503[Submit][S ...
- bzoj1264 [AHOI2006]基因匹配Match 树状数组+lcs
1264: [AHOI2006]基因匹配Match Time Limit: 10 Sec Memory Limit: 162 MBSubmit: 1255 Solved: 835[Submit][ ...
- BZOJ 1264: [AHOI2006]基因匹配Match 树状数组+DP
1264: [AHOI2006]基因匹配Match Description 基因匹配(match) 卡卡昨天晚上做梦梦见他和可可来到了另外一个星球,这个星球上生物的DNA序列由无数种碱基排列而成(地球 ...
- 【BZOJ1264】[AHOI2006]基因匹配Match DP+树状数组
[BZOJ1264][AHOI2006]基因匹配Match Description 基因匹配(match) 卡卡昨天晚上做梦梦见他和可可来到了另外一个星球,这个星球上生物的DNA序列由无数种碱基排列而 ...
- 基因匹配(bzoj 1264)
Description 基因匹配(match) 卡卡昨天晚上做梦梦见他和可可来到了另外一个星球,这个星球上生物的DNA序列由无数种碱基排列而成(地球上只有4种),而更奇怪的是,组成DNA序列的每一种碱 ...
- BZOJ1264: [AHOI2006]基因匹配Match
1264: [AHOI2006]基因匹配Match Time Limit: 10 Sec Memory Limit: 162 MBSubmit: 541 Solved: 347[Submit][S ...
- [AHOI2006]基因匹配
题目描述 卡卡昨天晚上做梦梦见他和可可来到了另外一个星球,这个星球上生物的DNA序列由无数种碱基排列而成(地球上只有4种),而更奇怪的是,组成DNA序列的每一种碱基在该序列中正好出现5次!这样如果一个 ...
- bzoj 1264: [AHOI2006]基因匹配Match
1264: [AHOI2006]基因匹配Match Description 基因匹配(match) 卡卡昨天晚上做梦梦见他和可可来到了另外一个星球,这个星球上生物的DNA序列由无数种碱基排列而成(地球 ...
随机推荐
- [转]基于VS Code快速搭建Java项目
有时候随手想写一点Java测试代码,以控制台程序为主,还会用到一些其它框架,并基于Maven构建. 1.Java Extension Pack一定要安装. 2.VS Code打开一个指定目录,创建相应 ...
- 4-3 xpath的用法
- Aizu 0531 "Paint Color" (坐标离散化+DFS or BFS)
传送门 题目描述: 为了宣传信息竞赛,要在长方形的三合板上喷油漆来制作招牌. 三合板上不需要涂色的部分预先贴好了护板. 被护板隔开的区域要涂上不同的颜色,比如上图就应该涂上5种颜色. 请编写一个程序计 ...
- 浅谈vue $mount()
Vue 的$mount()为手动挂载,在项目中可用于延时挂载(例如在挂载之前要进行一些其他操作.判断等),之后要手动挂载上.new Vue时,el和$mount并没有本质上的不同. 具体见代码: 顺便 ...
- The Zen of Python —— Python 之禅
Beautiful is better than ugly. # 优美好于丑陋(Python以编写优美的代码为目标) Explicit is better than implicit. # 明 ...
- IIS 6和IIS 7 中设置文件上传大小限制设置方法,两者是不一样的
在IIS 6.0中设置文件上传大小的方法,只要设置httpRuntime就可以了 <system.web> <httpRuntime executionTimeout="3 ...
- 23.logging
转载:https://www.cnblogs.com/yuanchenqi/article/5732581.html 一 (简单应用) import logging logging.debug('de ...
- 【算法随记七】巧用SIMD指令实现急速的字节流按位反转算法。
字节按位反转算法,在有些算法加密或者一些特殊的场合有着较为重要的应用,其速度也是一个非常关键的应用,比如一个byte变量a = 3,其二进制表示为00000011,进行按位反转后的结果即为110000 ...
- lombok工作原理分析
在Lombok使用的过程中,只需要添加相应的注解,无需再为此写任何代码.但是自动生成的代码到底是如何产生的呢? 核心之处就是对于注解的解析上.JDK5引入了注解的同时,也提供了两种解析方式. 运行时解 ...
- python获取网页信息的三种方法
import urllib.request import http.cookiejar url = 'http://www.baidu.com/' # 方法一 print('方法一') req_one ...
