吴裕雄--天生自然 PYTHON数据分析:基于Keras的CNN分析太空深处寻找系外行星数据
#We import libraries for linear algebra, graphs, and evaluation of results
import numpy as np
import matplotlib.pyplot as plt
from sklearn.linear_model import LinearRegression
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import roc_curve, roc_auc_score
from scipy.ndimage.filters import uniform_filter1d
#Keras is a high level neural networks library, based on either tensorflow or theano
from keras.models import Sequential, Model
from keras.layers import Conv1D, MaxPool1D, Dense, Dropout, Flatten, BatchNormalization, Input, concatenate, Activation
from keras.optimizers import Adam
INPUT_LIB = 'F:\\kaggleDataSet\\kepler-labelled\\'
raw_data = np.loadtxt(INPUT_LIB + 'exoTrain.csv', skiprows=1, delimiter=',')
x_train = raw_data[:, 1:]
y_train = raw_data[:, 0, np.newaxis] - 1.
raw_data = np.loadtxt(INPUT_LIB + 'exoTest.csv', skiprows=1, delimiter=',')
x_test = raw_data[:, 1:]
y_test = raw_data[:, 0, np.newaxis] - 1.
del raw_data
x_train = ((x_train - np.mean(x_train, axis=1).reshape(-1,1))/ np.std(x_train, axis=1).reshape(-1,1))
x_test = ((x_test - np.mean(x_test, axis=1).reshape(-1,1)) / np.std(x_test, axis=1).reshape(-1,1))
x_train = np.stack([x_train, uniform_filter1d(x_train, axis=1, size=200)], axis=2)
x_test = np.stack([x_test, uniform_filter1d(x_test, axis=1, size=200)], axis=2)
model = Sequential()
model.add(Conv1D(filters=8, kernel_size=11, activation='relu', input_shape=x_train.shape[1:]))
model.add(MaxPool1D(strides=4))
model.add(BatchNormalization())
model.add(Conv1D(filters=16, kernel_size=11, activation='relu'))
model.add(MaxPool1D(strides=4))
model.add(BatchNormalization())
model.add(Conv1D(filters=32, kernel_size=11, activation='relu'))
model.add(MaxPool1D(strides=4))
model.add(BatchNormalization())
model.add(Conv1D(filters=64, kernel_size=11, activation='relu'))
model.add(MaxPool1D(strides=4))
model.add(Flatten())
model.add(Dropout(0.5))
model.add(Dense(64, activation='relu'))
model.add(Dropout(0.25))
model.add(Dense(64, activation='relu'))
model.add(Dense(1, activation='sigmoid'))
def batch_generator(x_train, y_train, batch_size=32):
"""
Gives equal number of positive and negative samples, and rotates them randomly in time
"""
half_batch = batch_size // 2
x_batch = np.empty((batch_size, x_train.shape[1], x_train.shape[2]), dtype='float32')
y_batch = np.empty((batch_size, y_train.shape[1]), dtype='float32') yes_idx = np.where(y_train[:,0] == 1.)[0]
non_idx = np.where(y_train[:,0] == 0.)[0] while True:
np.random.shuffle(yes_idx)
np.random.shuffle(non_idx) x_batch[:half_batch] = x_train[yes_idx[:half_batch]]
x_batch[half_batch:] = x_train[non_idx[half_batch:batch_size]]
y_batch[:half_batch] = y_train[yes_idx[:half_batch]]
y_batch[half_batch:] = y_train[non_idx[half_batch:batch_size]] for i in range(batch_size):
sz = np.random.randint(x_batch.shape[1])
x_batch[i] = np.roll(x_batch[i], sz, axis = 0) yield x_batch, y_batch
#Start with a slightly lower learning rate, to ensure convergence
model.compile(optimizer=Adam(1e-5), loss = 'binary_crossentropy', metrics=['accuracy'])
hist = model.fit_generator(batch_generator(x_train, y_train, 32),
validation_data=(x_test, y_test),
verbose=0, epochs=5,
steps_per_epoch=x_train.shape[1]//32)
#Then speed things up a little
model.compile(optimizer=Adam(4e-5), loss = 'binary_crossentropy', metrics=['accuracy'])
hist = model.fit_generator(batch_generator(x_train, y_train, 32),
validation_data=(x_test, y_test),
verbose=2, epochs=40,
steps_per_epoch=x_train.shape[1]//32)


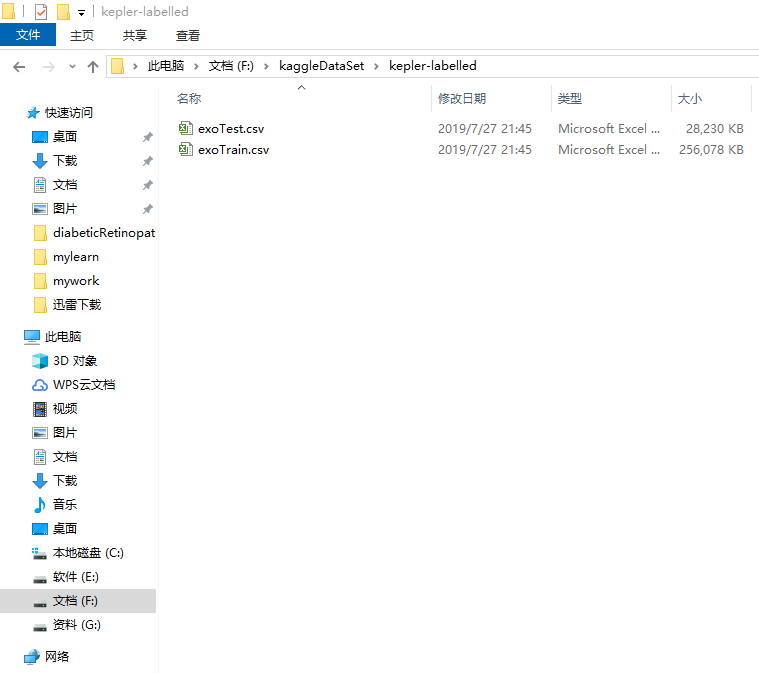
plt.plot(hist.history['loss'], color='b')
plt.plot(hist.history['val_loss'], color='r')
plt.show()
plt.plot(hist.history['acc'], color='b')
plt.plot(hist.history['val_acc'], color='r')
plt.show()
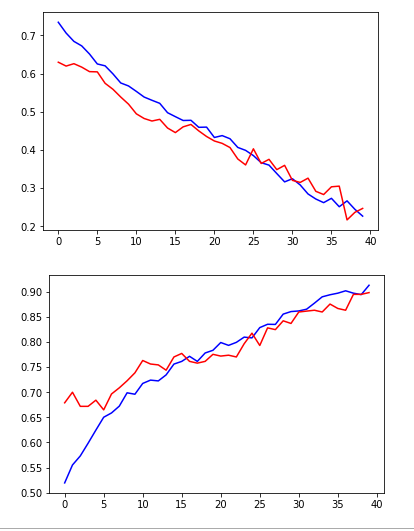
non_idx = np.where(y_test[:,0] == 0.)[0]
yes_idx = np.where(y_test[:,0] == 1.)[0]
y_hat = model.predict(x_test)[:,0]
plt.plot([y_hat[i] for i in yes_idx], 'bo')
plt.show()
plt.plot([y_hat[i] for i in non_idx], 'ro')
plt.show()
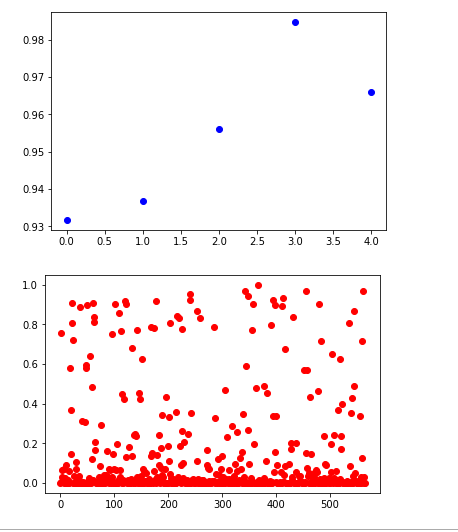
y_true = (y_test[:, 0] + 0.5).astype("int")
fpr, tpr, thresholds = roc_curve(y_true, y_hat)
plt.plot(thresholds, 1.-fpr)
plt.plot(thresholds, tpr)
plt.show()
crossover_index = np.min(np.where(1.-fpr <= tpr))
crossover_cutoff = thresholds[crossover_index]
crossover_specificity = 1.-fpr[crossover_index]
print("Crossover at {0:.2f} with specificity {1:.2f}".format(crossover_cutoff, crossover_specificity))
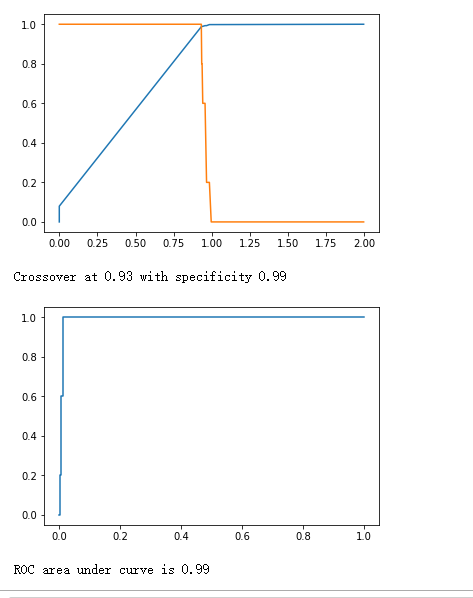
plt.plot(fpr, tpr)
plt.show()
print("ROC area under curve is {0:.2f}".format(roc_auc_score(y_true, y_hat)))
false_positives = np.where(y_hat * (1. - y_test) > 0.5)[0]
for i in non_idx:
if y_hat[i] > crossover_cutoff:
print(i)
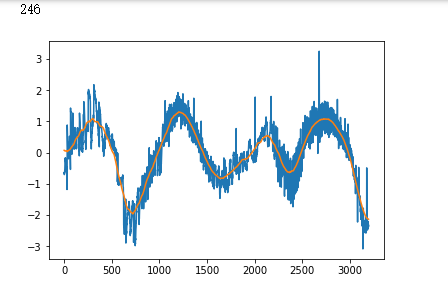
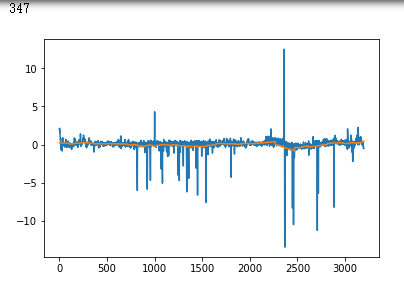
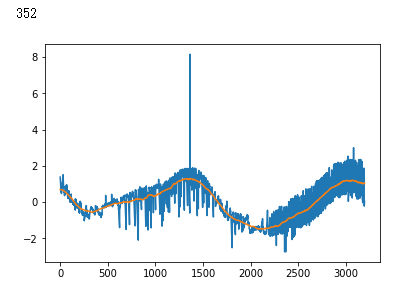
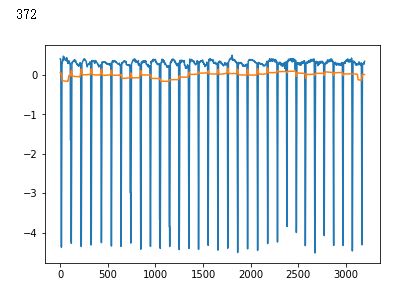
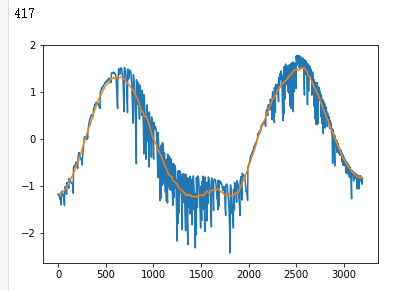
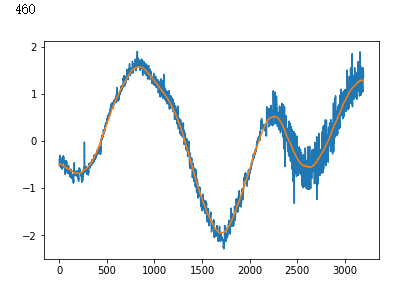
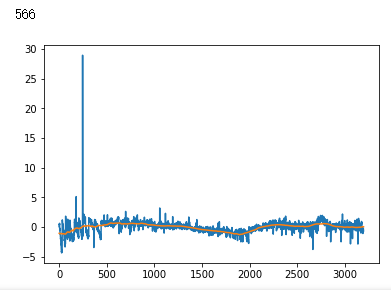
plt.plot(x_test[i])
plt.show()
吴裕雄--天生自然 PYTHON数据分析:基于Keras的CNN分析太空深处寻找系外行星数据的更多相关文章
- 吴裕雄--天生自然 python数据分析:健康指标聚集分析(健康分析)
# This Python 3 environment comes with many helpful analytics libraries installed # It is defined by ...
- 吴裕雄--天生自然 PYTHON数据分析:钦奈水资源管理分析
df = pd.read_csv("F:\\kaggleDataSet\\chennai-water\\chennai_reservoir_levels.csv") df[&quo ...
- 吴裕雄--天生自然 python数据分析:基于Keras使用CNN神经网络处理手写数据集
import pandas as pd import numpy as np import matplotlib.pyplot as plt import matplotlib.image as mp ...
- 吴裕雄--天生自然 PYTHON数据分析:糖尿病视网膜病变数据分析(完整版)
# This Python 3 environment comes with many helpful analytics libraries installed # It is defined by ...
- 吴裕雄--天生自然 PYTHON数据分析:所有美国股票和etf的历史日价格和成交量分析
# This Python 3 environment comes with many helpful analytics libraries installed # It is defined by ...
- 吴裕雄--天生自然 python数据分析:葡萄酒分析
# import pandas import pandas as pd # creating a DataFrame pd.DataFrame({'Yes': [50, 31], 'No': [101 ...
- 吴裕雄--天生自然 PYTHON数据分析:人类发展报告——HDI, GDI,健康,全球人口数据数据分析
import pandas as pd # Data analysis import numpy as np #Data analysis import seaborn as sns # Data v ...
- 吴裕雄--天生自然 python数据分析:医疗费数据分析
import numpy as np import pandas as pd import os import matplotlib.pyplot as pl import seaborn as sn ...
- 吴裕雄--天生自然 PYTHON数据分析:医疗数据分析
import numpy as np # linear algebra import pandas as pd # data processing, CSV file I/O (e.g. pd.rea ...
随机推荐
- UML-类图-需要写关联名称吗?
概念模型:需要写关联名称:类图:不需要写关联名称. 注意,概念模型关联线不需要箭头.
- linux 查看链接库的版本
我们编译可执行文件的时候,会链接各种依赖库, 但是怎么知道依赖库的版本正确呢? 下面有几种办法: ldd 这是比较差的,因为打印结果更与位置相关 dpkg -l | grep libprotobuf ...
- SLAM资料
当下SLAM方案的总体介绍 http://wwwbuild.net/roboteasy/908066.html slam基础知识 https://www.zhihu.com/question/3518 ...
- 编译x64c++出错,errorC1900:P1和P2之间 Il 不匹配问题
搜索了下相关资料,有一个说法是编译x64时本地缺失一些东西,2015安装update3就行. 我的是2013update4,找了下最新的有update5,安装然而并没有什么用. 最后还是重新找对应版本 ...
- mint 18中安装最新的R
mint 中默认的R版本有点老,升级最新版方法如下: 先卸载 sudo apt-get remove r-base-core 添加mint 18 识别的源 sudo echo "deb ht ...
- drf框架概况-resful接口规范-请求模块-渲染模块-Postman-drf请求生命周期
drf框架 全称:django-rest- framework 知识点: """ 1.接口:什么是接口.restful接口规范 2.CBV生命周期源码-基于restful ...
- ubuntu linux下解决“no java virtual machine was found after searching the following locations:”的方法
现象:删除旧的jdk,安装新的jdk之后,打开eclipse报错: A Java Runtime Environment (JRE) or Java Development Kit (JDK)must ...
- ios 接入微信开发 新版
首先在服务器所在域名(https://www.test.com)根目录创建apple-app-site-association文件 { "applinks": { "ap ...
- C实现日志等级控制
#include <stdio.h> #include <stdlib.h> #include <string.h> #include <stdarg.h&g ...
- rework-发出你的心声
生意人虚张声势的时候会给人什么感觉?都是些僵硬的措辞.官方的腔调.虚伪的友善.法律术语等.你一定看过这些玩意儿,就好像是机器人写出来的东西,这些公司在向你发话,而不是和你对话. 这种专业主义面具让人觉 ...
