CS231n 2016 通关 第三章-SVM 作业分析
作业内容,完成作业便可熟悉如下内容:
cell 1 设置绘图默认参数
# Run some setup code for this notebook. import random
import numpy as np
from cs231n.data_utils import load_CIFAR10
import matplotlib.pyplot as plt # This is a bit of magic to make matplotlib figures appear inline in the
# notebook rather than in a new window.
%matplotlib inline
plt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray' # Some more magic so that the notebook will reload external python modules;
# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython
%load_ext autoreload
%autoreload 2
cell 2 读取数据得到数组(矩阵)
# Load the raw CIFAR-10 data.
cifar10_dir = 'cs231n/datasets/cifar-10-batches-py'
X_train, y_train, X_test, y_test = load_CIFAR10(cifar10_dir) # As a sanity check, we print out the size of the training and test data.
print 'Training data shape: ', X_train.shape
print 'Training labels shape: ', y_train.shape
print 'Test data shape: ', X_test.shape
print 'Test labels shape: ', y_test.shape
得到结果,之后会用到,方便矩阵运算:

cell 3 对每一类随机选取对应的例子,并进行可视化:
# Visualize some examples from the dataset.
# We show a few examples of training images from each class.
classes = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']
num_classes = len(classes)
samples_per_class = 7
for y, cls in enumerate(classes):
idxs = np.flatnonzero(y_train == y)
idxs = np.random.choice(idxs, samples_per_class, replace=False)
for i, idx in enumerate(idxs):
plt_idx = i * num_classes + y + 1
plt.subplot(samples_per_class, num_classes, plt_idx)
plt.imshow(X_train[idx].astype('uint8'))
plt.axis('off')
if i == 0:
plt.title(cls)
plt.show()
可视化结果:

cell 4 将数据集划分为训练集,验证集。
# Split the data into train, val, and test sets. In addition we will
# create a small development set as a subset of the training data;
# we can use this for development so our code runs faster.
num_training = 49000
num_validation = 1000
num_test = 1000
num_dev = 500 # Our validation set will be num_validation points from the original
# training set.
mask = range(num_training, num_training + num_validation)
X_val = X_train[mask]
y_val = y_train[mask] # Our training set will be the first num_train points from the original
# training set.
mask = range(num_training)
X_train = X_train[mask]
y_train = y_train[mask] # We will also make a development set, which is a small subset of
# the training set.
mask = np.random.choice(num_training, num_dev, replace=False)
X_dev = X_train[mask]
y_dev = y_train[mask] # We use the first num_test points of the original test set as our
# test set.
mask = range(num_test)
X_test = X_test[mask]
y_test = y_test[mask] print 'Train data shape: ', X_train.shape
print 'Train labels shape: ', y_train.shape
print 'Validation data shape: ', X_val.shape
print 'Validation labels shape: ', y_val.shape
print 'Test data shape: ', X_test.shape
print 'Test labels shape: ', y_test.shape
得到各个数据集的尺寸:

cell 5 将数据reshape,方便计算
# Preprocessing: reshape the image data into rows
X_train = np.reshape(X_train, (X_train.shape[0], -1))
X_val = np.reshape(X_val, (X_val.shape[0], -1))
X_test = np.reshape(X_test, (X_test.shape[0], -1))
X_dev = np.reshape(X_dev, (X_dev.shape[0], -1)) # As a sanity check, print out the shapes of the data
print 'Training data shape: ', X_train.shape
print 'Validation data shape: ', X_val.shape
print 'Test data shape: ', X_test.shape
print 'dev data shape: ', X_dev.shape
reshape 后的结果

cell 6 计算训练集的均值
# Preprocessing: subtract the mean image
# first: compute the image mean based on the training data
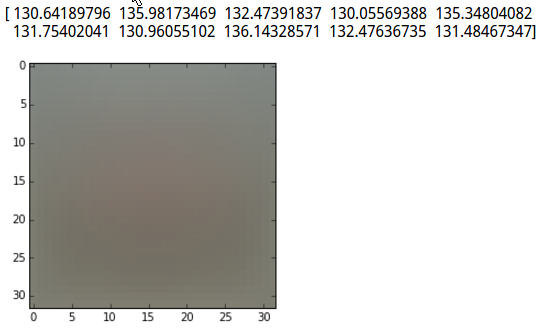
mean_image = np.mean(X_train, axis=0)
print mean_image[:10] # print a few of the elements
plt.figure(figsize=(4,4))
plt.imshow(mean_image.reshape((32,32,3)).astype('uint8')) # visualize the mean image
plt.show()
对均值可视化:
cell 7 训练集和测试集减去均值图像:
# second: subtract the mean image from train and test data
X_train -= mean_image
X_val -= mean_image
X_test -= mean_image
X_dev -= mean_image
cell 8 将截距项b添加到数据里,方便计算
# third: append the bias dimension of ones (i.e. bias trick) so that our SVM
# only has to worry about optimizing a single weight matrix W.
X_train = np.hstack([X_train, np.ones((X_train.shape[0], 1))])
X_val = np.hstack([X_val, np.ones((X_val.shape[0], 1))])
X_test = np.hstack([X_test, np.ones((X_test.shape[0], 1))])
X_dev = np.hstack([X_dev, np.ones((X_dev.shape[0], 1))]) print X_train.shape, X_val.shape, X_test.shape, X_dev.shape
数据尺寸:
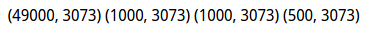
此前步骤为初始步骤,基本不需要修改,之后是作业核心。
cell 9 使用for循环计算loss
# Evaluate the naive implementation of the loss we provided for you:
from cs231n.classifiers.linear_svm import svm_loss_naive
import time # generate a random SVM weight matrix of small numbers
W = np.random.randn(3073, 10) * 0.0001 loss, grad = svm_loss_naive(W, X_dev, y_dev, 0.00001)
print 'loss: %f' % (loss, )
svm_loss_naive()具体实现:
def svm_loss_naive(W, X, y, reg):
"""
Structured SVM loss function, naive implementation (with loops). Inputs have dimension D, there are C classes, and we operate on minibatches
of N examples. Inputs:
- W: A numpy array of shape (D, C) containing weights.
- X: A numpy array of shape (N, D) containing a minibatch of data.
- y: A numpy array of shape (N,) containing training labels; y[i] = c means
that X[i] has label c, where 0 <= c < C.
- reg: (float) regularization strength Returns a tuple of:
- loss as single float
- gradient with respect to weights W; an array of same shape as W
"""
dW = np.zeros(W.shape) # initialize the gradient as zero # compute the loss and the gradient
# 10 classes
num_classes = W.shape[1]
#train 49000
num_train = X.shape[0]
loss = 0.0
"""
for i ->49000:
for j ->10:
compute every train smaple about each class
"""
#L = np.zeros(num_train,nun_class)
for i in xrange(num_train):
scores = X[i].dot(W)
#1*3073 * 3073*10 >>1*10
correct_class_score = scores[y[i]]
for j in xrange(num_classes):
if j == y[i]:
continue
margin = scores[j] - correct_class_score + 1 # note delta = 1
if margin > 0:
loss += margin
#dW 3073*10
dW[:,j] += X[i].T
dW[:,y[i]] -= X[i].T # Right now the loss is a sum over all training examples, but we want it
# to be an average instead so we divide by num_train.
loss /= num_train
dW /= num_train
# Add regularization to the loss.
# 1/2*w^2 w*w W*W = np.multiply(W,W) >>>>elementwise product
loss += reg * np.sum(W * W)
dW +=reg*W
#############################################################################
# TODO: #
# Compute the gradient of the loss function and store it dW. #
# Rather that first computing the loss and then computing the derivative, #
# it may be simpler to compute the derivative at the same time that the #
# loss is being computed. As a result you may need to modify some of the #
# code above to compute the gradient. #
#############################################################################
#computing coreect is differnt with others
return loss, dW
loss结果为8.97460
cell 10 完善svm_loss_naive(),梯度下降更新变量:
# Once you've implemented the gradient, recompute it with the code below
# and gradient check it with the function we provided for you # Compute the loss and its gradient at W.
loss, grad = svm_loss_naive(W, X_dev, y_dev, 0.0) # Numerically compute the gradient along several randomly chosen dimensions, and
# compare them with your analytically computed gradient. The numbers should match
# almost exactly along all dimensions.
from cs231n.gradient_check import grad_check_sparse
f = lambda w: svm_loss_naive(w, X_dev, y_dev, 0.0)[0]
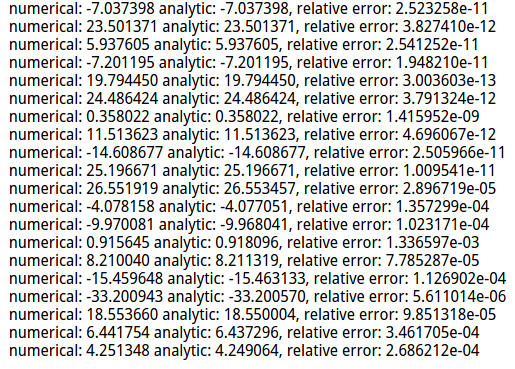
grad_numerical = grad_check_sparse(f, W, grad) # do the gradient check once again with regularization turned on
# you didn't forget the regularization gradient did you?
loss, grad = svm_loss_naive(W, X_dev, y_dev, 1e2)
f = lambda w: svm_loss_naive(w, X_dev, y_dev, 1e2)[0]
grad_numerical = grad_check_sparse(f, W, grad)
梯度更新代码已在cell9调用时给出。
梯度校验》》》使用解析法得到的dw与数值计算法得到的结果比较:
问题:

cell 11 使用向量(矩阵)的方式计算loss,并与之前的结果比较:
# Next implement the function svm_loss_vectorized; for now only compute the loss;
# we will implement the gradient in a moment.
tic = time.time()
loss_naive, grad_naive = svm_loss_naive(W, X_dev, y_dev, 0.00001)
toc = time.time()
print 'Naive loss: %e computed in %fs' % (loss_naive, toc - tic) from cs231n.classifiers.linear_svm import svm_loss_vectorized
tic = time.time()
loss_vectorized, _ = svm_loss_vectorized(W, X_dev, y_dev, 0.00001)
toc = time.time()
print 'Vectorized loss: %e computed in %fs' % (loss_vectorized, toc - tic) # The losses should match but your vectorized implementation should be much faster.
print 'difference: %f' % (loss_naive - loss_vectorized)
差异结果:
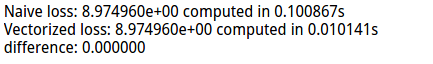
cell 12 使用向量(矩阵)的方式计算grad,并与之前的结果比较:
# Complete the implementation of svm_loss_vectorized, and compute the gradient
# of the loss function in a vectorized way. # The naive implementation and the vectorized implementation should match, but
# the vectorized version should still be much faster.
tic = time.time()
_, grad_naive = svm_loss_naive(W, X_dev, y_dev, 0.00001)
toc = time.time()
print 'Naive loss and gradient: computed in %fs' % (toc - tic) tic = time.time()
_, grad_vectorized = svm_loss_vectorized(W, X_dev, y_dev, 0.00001)
toc = time.time()
print 'Vectorized loss and gradient: computed in %fs' % (toc - tic) # The loss is a single number, so it is easy to compare the values computed
# by the two implementations. The gradient on the other hand is a matrix, so
# we use the Frobenius norm to compare them.
difference = np.linalg.norm(grad_naive - grad_vectorized, ord='fro')
print 'difference: %f' % difference
结果比较:

cell 13 随机梯度下降》》SGD
# In the file linear_classifier.py, implement SGD in the function
# LinearClassifier.train() and then run it with the code below.
from cs231n.classifiers import LinearSVM
svm = LinearSVM()
tic = time.time()
loss_hist = svm.train(X_train, y_train, learning_rate=1e-7, reg=5e4,
num_iters=1500, verbose=True)
toc = time.time()
print 'That took %fs' % (toc - tic)
loss 结果:

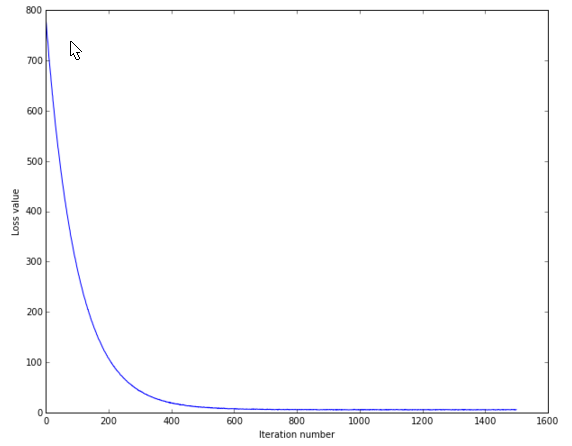
cell 14 画出loss变化曲线:
# A useful debugging strategy is to plot the loss as a function of
# iteration number:
plt.plot(loss_hist)
plt.xlabel('Iteration number')
plt.ylabel('Loss value')
plt.show()
结果:
cell 15 用上述训练的参数,对验证集与训练集进行测试:
# Write the LinearSVM.predict function and evaluate the performance on both the
# training and validation set
y_train_pred = svm.predict(X_train)
print 'training accuracy: %f' % (np.mean(y_train == y_train_pred), )
y_val_pred = svm.predict(X_val)
print 'validation accuracy: %f' % (np.mean(y_val == y_val_pred), )
结果:

cell 16 对超参数》》学习率 正则化强度 使用训练集进行训练,使用验证集验证结果,选取最好的超参数。
# Use the validation set to tune hyperparameters (regularization strength and
# learning rate). You should experiment with different ranges for the learning
# rates and regularization strengths; if you are careful you should be able to
# get a classification accuracy of about 0.4 on the validation set.
learning_rates = [1e-7, 2e-7, 3e-7, 5e-5, 8e-7]
regularization_strengths = [1e4, 2e4, 3e4, 4e4, 5e4, 6e4, 7e4, 8e4, 1e5] # results is dictionary mapping tuples of the form
# (learning_rate, regularization_strength) to tuples of the form
# (training_accuracy, validation_accuracy). The accuracy is simply the fraction
# of data points that are correctly classified.
results = {}
best_val = -1 # The highest validation accuracy that we have seen so far.
best_svm = None # The LinearSVM object that achieved the highest validation rate. ################################################################################
# TODO: #
# Write code that chooses the best hyperparameters by tuning on the validation #
# set. For each combination of hyperparameters, train a linear SVM on the #
# training set, compute its accuracy on the training and validation sets, and #
# store these numbers in the results dictionary. In addition, store the best #
# validation accuracy in best_val and the LinearSVM object that achieves this #
# accuracy in best_svm. #
# #
# Hint: You should use a small value for num_iters as you develop your #
# validation code so that the SVMs don't take much time to train; once you are #
# confident that your validation code works, you should rerun the validation #
# code with a larger value for num_iters. #
################################################################################
iters = 1500
for lr in learning_rates:
for rs in regularization_strengths:
svm = LinearSVM()
svm.train(X_train, y_train, learning_rate=lr, reg=rs, num_iters=iters)
y_train_pred = svm.predict(X_train)
accu_train = np.mean(y_train == y_train_pred)
y_val_pred = svm.predict(X_val)
accu_val = np.mean(y_val == y_val_pred) results[(lr, rs)] = (accu_train, accu_val) if best_val < accu_val:
best_val = accu_val
best_svm = svm
################################################################################
# END OF YOUR CODE #
################################################################################ # Print out results.
for lr, reg in sorted(results):
train_accuracy, val_accuracy = results[(lr, reg)]
print 'lr %e reg %e train accuracy: %f val accuracy: %f' % (
lr, reg, train_accuracy, val_accuracy) print 'best validation accuracy achieved during cross-validation: %f' % best_val
最终结果:

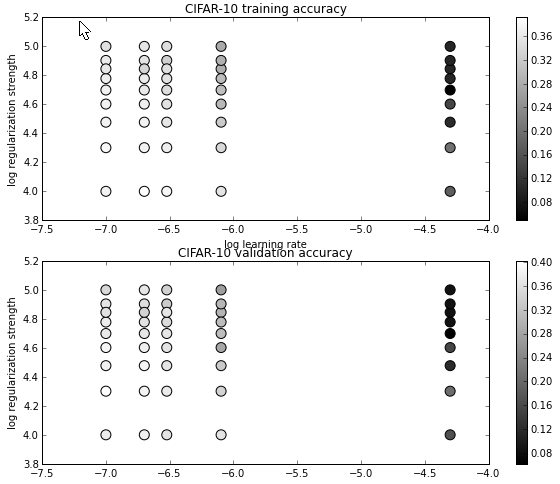
cell 17 对各组超参数的结果可视化:
# Visualize the cross-validation results
import math
x_scatter = [math.log10(x[0]) for x in results]
y_scatter = [math.log10(x[1]) for x in results] # plot training accuracy
marker_size = 100
colors = [results[x][0] for x in results]
plt.subplot(2, 1, 1)
plt.scatter(x_scatter, y_scatter, marker_size, c=colors)
plt.colorbar()
plt.xlabel('log learning rate')
plt.ylabel('log regularization strength')
plt.title('CIFAR-10 training accuracy') # plot validation accuracy
colors = [results[x][1] for x in results] # default size of markers is 20
plt.subplot(2, 1, 2)
plt.scatter(x_scatter, y_scatter, marker_size, c=colors)
plt.colorbar()
plt.xlabel('log learning rate')
plt.ylabel('log regularization strength')
plt.title('CIFAR-10 validation accuracy')
plt.show()
显示结果:
cell 18 使用得到最好超参数的模型对test集进行预测,比较预测值与真实值,计算准确率。
# Evaluate the best svm on test set
y_test_pred = best_svm.predict(X_test)
test_accuracy = np.mean(y_test == y_test_pred)
print 'linear SVM on raw pixels final test set accuracy: %f' % test_accuracy
结果:

cell 19 对得到的w对应的class进行可视化:
# Visualize the learned weights for each class.
# Depending on your choice of learning rate and regularization strength, these may
# or may not be nice to look at.
w = best_svm.W[:-1,:] # strip out the bias
w = w.reshape(32, 32, 3, 10)
w_min, w_max = np.min(w), np.max(w)
classes = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']
for i in xrange(10):
plt.subplot(2, 5, i + 1) # Rescale the weights to be between 0 and 255
wimg = 255.0 * (w[:, :, :, i].squeeze() - w_min) / (w_max - w_min)
plt.imshow(wimg.astype('uint8'))
plt.axis('off')
plt.title(classes[i])
结果:

问题:

注:在softmax 与svm的超参数选择时,使用了共同的类,以及类中不同的相应的方法。具体的文件内容与注释如下。
附:通关CS231n企鹅群:578975100 validation:DL-CS231n
CS231n 2016 通关 第三章-SVM 作业分析的更多相关文章
- CS231n 2016 通关 第三章-Softmax 作业
在完成SVM作业的基础上,Softmax的作业相对比较轻松. 完成本作业需要熟悉与掌握的知识: cell 1 设置绘图默认参数 mport random import numpy as np from ...
- CS231n 2016 通关 第三章-SVM与Softmax
1===本节课对应视频内容的第三讲,对应PPT是Lecture3 2===本节课的收获 ===熟悉SVM及其多分类问题 ===熟悉softmax分类问题 ===了解优化思想 由上节课即KNN的分析步骤 ...
- CS231n 2016 通关 第四章-NN 作业
cell 1 显示设置初始化 # A bit of setup import numpy as np import matplotlib.pyplot as plt from cs231n.class ...
- CS231n 2016 通关 第五章 Training NN Part1
在上一次总结中,总结了NN的基本结构. 接下来的几次课,对一些具体细节进行讲解. 比如激活函数.参数初始化.参数更新等等. ====================================== ...
- CS231n 2016 通关 第六章 Training NN Part2
本章节讲解 参数更新 dropout ================================================================================= ...
- CS231n 2016 通关 第四章-反向传播与神经网络(第一部分)
在上次的分享中,介绍了模型建立与使用梯度下降法优化参数.梯度校验,以及一些超参数的经验. 本节课的主要内容: 1==链式法则 2==深度学习框架中链式法则 3==全连接神经网络 =========== ...
- CSAPP深入理解计算机系统(第二版)第三章家庭作业答案
<深入理解计算机系统(第二版)>CSAPP 第三章 家庭作业 这一章介绍了AT&T的汇编指令 比较重要 本人完成了<深入理解计算机系统(第二版)>(以下简称CSAPP) ...
- C++第三章课后作业答案及解析---指针的使用
今天继续完成上周没有完成的习题---C++第三章课后作业,本章题涉及指针的使用,有指向对象的指针做函数参数,对象的引用以及友元类的使用方法等 它们具体的使用方法在下面的题目中会有具体的解析(解析标注在 ...
- Hand on Machine Learning第三章课后作业(1):垃圾邮件分类
import os import email import email.policy 1. 读取邮件数据 SPAM_PATH = os.path.join( "E:\\3.Study\\机器 ...
随机推荐
- vue-class-component 以class的模式写vue组件
vue英文官网推荐了一个叫vue-class-component的包,可以以class的模式写vue组件.vue-class-component(以下简称Component)带来了很多便利: 1.me ...
- overflow滚动条样式设置,ie和webkit内核
<!DOCTYPE html> <html lang="en"> <head> <meta charset="UTF-8&quo ...
- matplotlib简易新手教程及动画
做数据分析,首先是要熟悉和理解数据.所以掌握一个趁手的可视化工具是很重要的,否则对数据连个主要的感性认识都没有,怎样进行下一步的design 点击打开链接 还有一个非常棒的资料 Matplotlib ...
- 纯JS设置首页,增加收藏,获取URL參数,解决中文乱码
雪影工作室版权全部,转载请注明[http://blog.csdn.net/lina791211] 1.前言 纯Javascript 设置首页,增加收藏. 2.设置首页 // 设置为主页 functio ...
- HDU 3435A new Graph Game(网络流之最小费用流)
题目地址:HDU 3435 这题刚上来一看,感觉毫无头绪. .再细致想想.. 发现跟我做的前两道费用流的题是差点儿相同的. 能够往那上面转换. 建图基本差点儿相同.仅仅只是这里是无向图.建图依旧是拆点 ...
- solaris软件管理 FTP
安装一些常用软件 一.应用程序与系统命令的关系: 系统命令文件位置在 /bin /sbin下面或为shell内部指令:完成对系统的基本管理工作:一般在字符操作界面中运行:一般包括命令字.命令选项和命令 ...
- selector的button选中处理问题
1.背景介绍 在做Android项目开发的时候,有时我们须要对button做一些特殊的处理,比方button点击的时候会有一个动画的效果,实际上就是几张图片在短时间的切换.再比方有时候我们须要对界面的 ...
- uboot下载地址
非常奇怪百度搜索都搜不到一个好的uboot下载的说明,仅此标记 HOME http://www.denx.de/wiki/U-Boot/SourceCode sourcecode http://www ...
- 04 http协议模拟登陆发帖
<?php require('./http.class.php'); $http = new Http('http://home.verycd.com/cp.php?ac=pm&op=s ...
- Linux就该这么学--命令集合7(管道命令符)
1.管道命令符“|”的作用是将前一个命令的标准输出当作后一个命令的标准输入,格式为:“命令A|命令B”. 找出被限制登录用户的命令是:grep "/sbin/nologin" /e ...
