吴裕雄--天生自然 R语言开发学习:分类














#-----------------------------------------------------------------------------#
# R in Action (2nd ed): Chapter 17 #
# Classification #
# requires packaged rpart, party, randomForest, kernlab, rattle #
# install.packages(c("rpart", "party", "randomForest", "e1071", "rpart.plot") #
# install.packages(rattle, dependencies = c("Depends", "Suggests")) #
#-----------------------------------------------------------------------------# par(ask=TRUE) # Listing 17.1 - Prepare the breast cancer data
loc <- "http://archive.ics.uci.edu/ml/machine-learning-databases/"
ds <- "breast-cancer-wisconsin/breast-cancer-wisconsin.data"
url <- paste(loc, ds, sep="") breast <- read.table(url, sep=",", header=FALSE, na.strings="?")
names(breast) <- c("ID", "clumpThickness", "sizeUniformity",
"shapeUniformity", "maginalAdhesion",
"singleEpithelialCellSize", "bareNuclei",
"blandChromatin", "normalNucleoli", "mitosis", "class") df <- breast[-1]
df$class <- factor(df$class, levels=c(2,4),
labels=c("benign", "malignant")) set.seed(1234)
train <- sample(nrow(df), 0.7*nrow(df))
df.train <- df[train,]
df.validate <- df[-train,]
table(df.train$class)
table(df.validate$class) # Listing 17.2 - Logistic regression with glm()
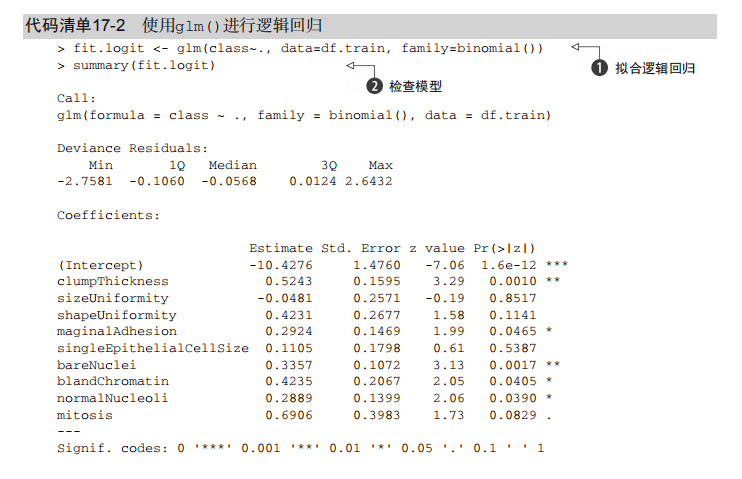
fit.logit <- glm(class~., data=df.train, family=binomial())
summary(fit.logit)
prob <- predict(fit.logit, df.validate, type="response")
logit.pred <- factor(prob > .5, levels=c(FALSE, TRUE),
labels=c("benign", "malignant"))
logit.perf <- table(df.validate$class, logit.pred,
dnn=c("Actual", "Predicted"))
logit.perf # Listing 17.3 - Creating a classical decision tree with rpart()
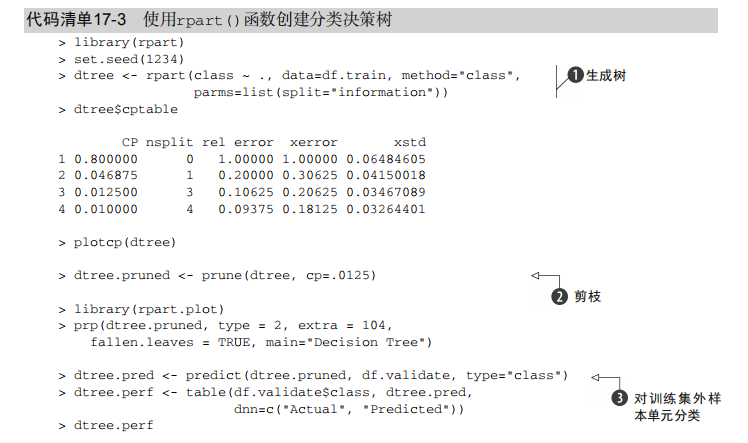
library(rpart)
set.seed(1234)
dtree <- rpart(class ~ ., data=df.train, method="class",
parms=list(split="information"))
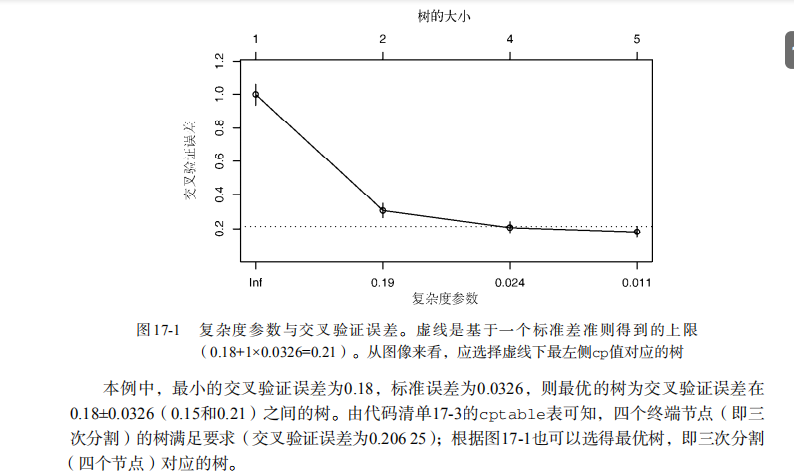
dtree$cptable
plotcp(dtree) dtree.pruned <- prune(dtree, cp=.0125) library(rpart.plot)
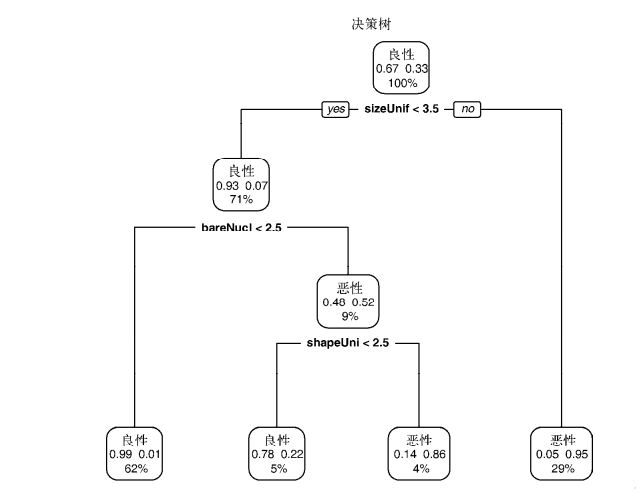
prp(dtree.pruned, type = 2, extra = 104,
fallen.leaves = TRUE, main="Decision Tree") dtree.pred <- predict(dtree.pruned, df.validate, type="class")
dtree.perf <- table(df.validate$class, dtree.pred,
dnn=c("Actual", "Predicted"))
dtree.perf # Listing 17.4 - Creating a conditional inference tree with ctree()
library(party)
fit.ctree <- ctree(class~., data=df.train)
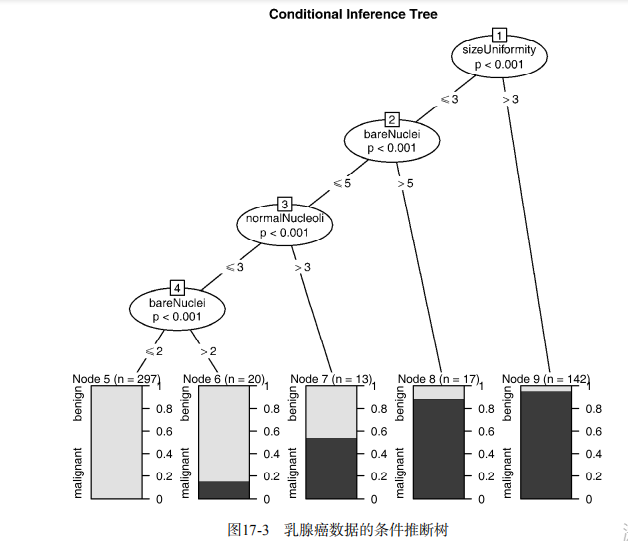
plot(fit.ctree, main="Conditional Inference Tree") ctree.pred <- predict(fit.ctree, df.validate, type="response")
ctree.perf <- table(df.validate$class, ctree.pred,
dnn=c("Actual", "Predicted"))
ctree.perf # Listing 17.5 - Random forest
library(randomForest)
set.seed(1234)
fit.forest <- randomForest(class~., data=df.train,
na.action=na.roughfix,
importance=TRUE)
fit.forest
importance(fit.forest, type=2)
forest.pred <- predict(fit.forest, df.validate)
forest.perf <- table(df.validate$class, forest.pred,
dnn=c("Actual", "Predicted"))
forest.perf # Listing 17.6 - A support vector machine
library(e1071)
set.seed(1234)
fit.svm <- svm(class~., data=df.train)
fit.svm
svm.pred <- predict(fit.svm, na.omit(df.validate))
svm.perf <- table(na.omit(df.validate)$class,
svm.pred, dnn=c("Actual", "Predicted"))
svm.perf # Listing 17.7 Tuning an RBF support vector machine (this can take a while)
set.seed(1234)
tuned <- tune.svm(class~., data=df.train,
gamma=10^(-6:1),
cost=10^(-10:10))
tuned
fit.svm <- svm(class~., data=df.train, gamma=.01, cost=1)
svm.pred <- predict(fit.svm, na.omit(df.validate))
svm.perf <- table(na.omit(df.validate)$class,
svm.pred, dnn=c("Actual", "Predicted"))
svm.perf # Listing 17.8 Function for assessing binary classification accuracy
performance <- function(table, n=2){
if(!all(dim(table) == c(2,2)))
stop("Must be a 2 x 2 table")
tn = table[1,1]
fp = table[1,2]
fn = table[2,1]
tp = table[2,2]
sensitivity = tp/(tp+fn)
specificity = tn/(tn+fp)
ppp = tp/(tp+fp)
npp = tn/(tn+fn)
hitrate = (tp+tn)/(tp+tn+fp+fn)
result <- paste("Sensitivity = ", round(sensitivity, n) ,
"\nSpecificity = ", round(specificity, n),
"\nPositive Predictive Value = ", round(ppp, n),
"\nNegative Predictive Value = ", round(npp, n),
"\nAccuracy = ", round(hitrate, n), "\n", sep="")
cat(result)
} # Listing 17.9 - Performance of breast cancer data classifiers
performance(dtree.perf)
performance(ctree.perf)
performance(forest.perf)
performance(svm.perf) # Using Rattle Package for data mining loc <- "http://archive.ics.uci.edu/ml/machine-learning-databases/"
ds <- "pima-indians-diabetes/pima-indians-diabetes.data"
url <- paste(loc, ds, sep="")
diabetes <- read.table(url, sep=",", header=FALSE)
names(diabetes) <- c("npregant", "plasma", "bp", "triceps",
"insulin", "bmi", "pedigree", "age", "class")
diabetes$class <- factor(diabetes$class, levels=c(0,1),
labels=c("normal", "diabetic"))
library(rattle)
rattle()
吴裕雄--天生自然 R语言开发学习:分类的更多相关文章
- 吴裕雄--天生自然 R语言开发学习:R语言的安装与配置
下载R语言和开发工具RStudio安装包 先安装R
- 吴裕雄--天生自然 R语言开发学习:数据集和数据结构
数据集的概念 数据集通常是由数据构成的一个矩形数组,行表示观测,列表示变量.表2-1提供了一个假想的病例数据集. 不同的行业对于数据集的行和列叫法不同.统计学家称它们为观测(observation)和 ...
- 吴裕雄--天生自然 R语言开发学习:导入数据
2.3.6 导入 SPSS 数据 IBM SPSS数据集可以通过foreign包中的函数read.spss()导入到R中,也可以使用Hmisc 包中的spss.get()函数.函数spss.get() ...
- 吴裕雄--天生自然 R语言开发学习:使用键盘、带分隔符的文本文件输入数据
R可从键盘.文本文件.Microsoft Excel和Access.流行的统计软件.特殊格 式的文件.多种关系型数据库管理系统.专业数据库.网站和在线服务中导入数据. 使用键盘了.有两种常见的方式:用 ...
- 吴裕雄--天生自然 R语言开发学习:R语言的简单介绍和使用
假设我们正在研究生理发育问 题,并收集了10名婴儿在出生后一年内的月龄和体重数据(见表1-).我们感兴趣的是体重的分 布及体重和月龄的关系. 可以使用函数c()以向量的形式输入月龄和体重数据,此函 数 ...
- 吴裕雄--天生自然 R语言开发学习:基础知识
1.基础数据结构 1.1 向量 # 创建向量a a <- c(1,2,3) print(a) 1.2 矩阵 #创建矩阵 mymat <- matrix(c(1:10), nrow=2, n ...
- 吴裕雄--天生自然 R语言开发学习:图形初阶(续二)
# ----------------------------------------------------# # R in Action (2nd ed): Chapter 3 # # Gettin ...
- 吴裕雄--天生自然 R语言开发学习:图形初阶(续一)
# ----------------------------------------------------# # R in Action (2nd ed): Chapter 3 # # Gettin ...
- 吴裕雄--天生自然 R语言开发学习:图形初阶
# ----------------------------------------------------# # R in Action (2nd ed): Chapter 3 # # Gettin ...
- 吴裕雄--天生自然 R语言开发学习:基本图形(续二)
#---------------------------------------------------------------# # R in Action (2nd ed): Chapter 6 ...
随机推荐
- 面向对象 part6 继承
继承 js实现的是实现继承/也就是继承实际的方法 //主要依赖:原型链 //基本思路: 就是一个引用类型继承另一个引用类型的属性和方法 详细:构造函数,实例,原型之间的关系.每个构造函数都有一个原型对 ...
- JavaSE--jdom解析之bom
参考:http://www.cnblogs.com/findumars/p/3620078.html org.jdom2.input.JDOMParseException: Error on line ...
- Angular(三)
Angular开发者指南(三)数据绑定 数据绑定AngularJS应用程序中的数据绑定是模型和视图组件之间的数据的自动同步. AngularJS实现数据绑定的方式可以将模型视为应用程序中的单一来源 ...
- 01 语言基础+高级:1-4 接口与多态_day10【接口、多态】&&day11【final、匿名内部类】
day10[接口.多态] 接口三大特征——多态引用类型转换 教学目标写出定义接口的格式写出实现接口的格式说出接口中成员的特点能够说出使用多态的前提条件理解多态的向上转型理解多态的向下转型 day10_ ...
- Python访问Amazon官网异常
使用Python访问亚马逊(Amazon)官网,如果没有将headers更改为浏览器的信息, 有几率会触发:检测到当前可能是自动程序,需要输入验证码: 将header修改成浏览器后,需要等一段时间或者 ...
- HTTP编码
HTTP编码 不仅仅URL需要编码,HTTP header也需要编码,HTTP body 无特殊要求 一般采用百分号编码:比如一个字节的ascii码值是 0x89 那使用百分号编码之后 输出是 %89 ...
- Kafa 的安装配置及使用
1.kafka 的简介及应用场景 Apache Kafka是一个分布式的消息系统,可用于统计,日志及流处理 2.kafka 基本原理 3.kafka 集群体系结构 4.kafka实例 https:// ...
- EternalBlue永恒之蓝漏洞复现
EternalBlue漏洞复现 1. 实训目的 永恒之蓝(EternalBlue)是由美国国家安全局开发的漏洞利用程序,对应微软漏洞编号ms17-010.该漏洞利用工具由一个名为”影子经济人”( ...
- 求最近公共祖先(LCA)的各种算法
水一发题解. 我只是想存一下树剖LCA的代码...... 以洛谷上的这个模板为例:P3379 [模板]最近公共祖先(LCA) 1.朴素LCA 就像做模拟题一样,先dfs找到基本信息:每个节点的父亲.深 ...
- dao层单元测试报错CONDITIONS EVALUATION REPORT
0 环境 系统:win10 编辑器:IDEA 1 正文 1.1 起因 在controller层测试 测试url时没问题的 但是我单元测试就报错 1.2 排查 因为controller层 springb ...
