HDU1560 DNA sequence —— IDA*算法
题目链接:http://acm.hdu.edu.cn/showproblem.php?pid=1560
DNA sequence
Time Limit: 15000/5000 MS (Java/Others) Memory Limit: 32768/32768 K (Java/Others)
Total Submission(s): 2999 Accepted Submission(s): 1462
sequences is one of the basic problems in modern computational molecular biology. But this problem is a little different. Given several DNA sequences, you are asked to make a shortest sequence from them so that each of the given sequence is the subsequence
of it.
For example, given "ACGT","ATGC","CGTT" and "CAGT", you can make a sequence in the following way. It is the shortest but may be not the only one.
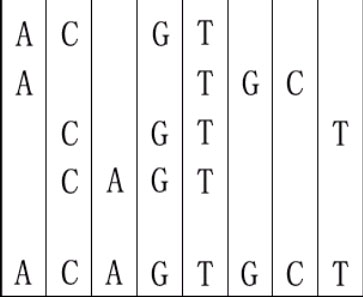
sequence is between 1 and 5.
4
ACGT
ATGC
CGTT
CAGT
题解:
一开始以为是直接用回溯的方法,结果TLE。看了题解是用IDA*(迭代加深搜),其实自己不太了解迭代加深搜为什么比较快,而且什么时候用合适?下面是自己对迭代加深搜的一些浅薄的了解:
1.首先迭代加深搜适合用在:求最少步数(带有BFS的特点)并且不太容易估计搜索深度的问题上,同时兼有了BFS求最少步数和DFS易写、无需多开数组的特点。
2.相对于赤裸裸的回溯,迭代加深搜由于限制了搜索深度,所以也能适当地剪枝。
3.我编不下去了……
代码一:
#include <iostream>
#include <cstdio>
#include <cstring>
#include <cmath>
#include <algorithm>
#include <vector>
#include <queue>
#include <stack>
#include <map>
#include <string>
#include <set>
#define ms(a,b) memset((a),(b),sizeof((a)))
using namespace std;
typedef long long LL;
const int INF = 2e9;
const LL LNF = 9e18;
const int MOD = 1e9+;
const int MAXN = +; int n;
char dna[MAXN][MAXN];
int len[MAXN], pos[MAXN];
char s[] = {'A', 'G', 'C', 'T'}; bool dfs(int k, int limit) //k为放了几个, k+1才为当前要放的
{
int maxx = , cnt = ; //maxx为最长剩余的dna片段, cnt为剩余的片段之和(核苷酸链?好怀念啊)
for(int i = ; i<n; i++)
{
cnt += len[i]-pos[i];
maxx = max(maxx, len[i]-pos[i]);
}
if(cnt==) return true; //如果片段都放完,则已得到答案
if(cnt<=limit-k) return true; //剪枝:片段之和小于等于剩余能放数量,肯定能够得到答案
if(maxx>limit-k) return false; //剪枝:最小的估计值都大于剩余能放数量,肯定不能得到答案 int tmp[MAXN];
for(int i = ; i<; i++)
{
memcpy(tmp, pos, sizeof(tmp));
bool flag = false;
for(int j = ; j<n; j++)
if(dna[j][pos[j]]==s[i])
pos[j]++, flag = true; //k+1<=limit:在限制范围内
if(k+<=limit && flag && dfs(k+, limit) )
return true;
memcpy(pos, tmp, sizeof(pos));
}
return false;
} int main()
{
int T;
scanf("%d",&T);
while(T--)
{
scanf("%d",&n);
int limit = ;
for(int i = ; i<n; i++)
{
scanf("%s",dna[i]);
len[i] = strlen(dna[i]);
limit = max(limit, len[i]);
} ms(pos, );
while(!dfs(, limit))
limit++;
printf("%d\n", limit);
}
}
代码二:
#include <iostream>
#include <cstdio>
#include <cstring>
#include <cmath>
#include <algorithm>
#include <vector>
#include <queue>
#include <stack>
#include <map>
#include <string>
#include <set>
#define ms(a,b) memset((a),(b),sizeof((a)))
using namespace std;
typedef long long LL;
const int INF = 2e9;
const LL LNF = 9e18;
const int MOD = 1e9+;
const int MAXN = +; int n;
char dna[MAXN][MAXN];
int len[MAXN], pos[MAXN];
char s[] = {'A', 'G', 'C', 'T'}; bool dfs(int k, int limit) //k为放了几个, k+1才为当前要放的
{
if(k>limit) return false; int maxx = , cnt = ; //maxx为最长剩余的dna片段, cnt为剩余的片段之和(核苷酸链?好怀念啊)
for(int i = ; i<n; i++)
{
cnt += len[i]-pos[i];
maxx = max(maxx, len[i]-pos[i]);
}
if(cnt==) return true; //如果片段都放完,则已得到答案
if(cnt<=limit-k) return true; //剪枝:片段之和小于等于剩余能放数量,肯定能够得到答案
if(maxx>limit-k) return false; //剪枝:最小的估计值都大于剩余能放数量,肯定不能得到答案 int tmp[MAXN];
for(int i = ; i<; i++)
{
memcpy(tmp, pos, sizeof(tmp));
bool flag = false;
for(int j = ; j<n; j++)
if(dna[j][pos[j]]==s[i])
pos[j]++, flag = true; if(flag && dfs(k+, limit) )
return true;
memcpy(pos, tmp, sizeof(pos));
}
return false;
} int main()
{
int T;
scanf("%d",&T);
while(T--)
{
scanf("%d",&n);
int limit = ;
for(int i = ; i<n; i++)
{
scanf("%s",dna[i]);
len[i] = strlen(dna[i]);
limit = max(limit, len[i]);
} ms(pos, );
while(!dfs(, limit))
limit++;
printf("%d\n", limit);
}
}
HDU1560 DNA sequence —— IDA*算法的更多相关文章
- HDU1560 DNA sequence IDA* + 强力剪枝 [kuangbin带你飞]专题二
题意:给定一些DNA序列,求一个最短序列能够包含所有序列. 思路:记录第i个序列已经被匹配的长度p[i],以及第i序列的原始长度len[i].则有两个剪枝: 剪枝1:直接取最长待匹配长度.1900ms ...
- HDU1560 DNA sequence(IDA*)题解
DNA sequence Time Limit: 15000/5000 MS (Java/Others) Memory Limit: 32768/32768 K (Java/Others) To ...
- Hdu1560 DNA sequence(IDA*) 2017-01-20 18:53 50人阅读 评论(0) 收藏
DNA sequence Time Limit : 15000/5000ms (Java/Other) Memory Limit : 32768/32768K (Java/Other) Total ...
- HDU1560 DNA sequence
题目: The twenty-first century is a biology-technology developing century. We know that a gene is made ...
- HDU 1560 DNA sequence (IDA* 迭代加深 搜索)
题目地址:http://acm.hdu.edu.cn/showproblem.php?pid=1560 BFS题解:http://www.cnblogs.com/crazyapple/p/321810 ...
- HDU 1560 DNA sequence(IDA*)
题目链接:http://acm.hdu.edu.cn/showproblem.php?pid=1560 题目大意:给出n个字符串,让你找一个字符串使得这n个字符串都是它的子串,求最小长度. 解题思路: ...
- DNA sequence HDU - 1560(IDA*,迭代加深搜索)
题目大意:有n个DNA序列,构造一个新的序列,使得这n个DNA序列都是它的子序列,然后输出最小长度. 题解:第一次接触IDA*算法,感觉~~好暴力!!思路:维护一个数组pos[i],表示第i个串该匹配 ...
- 【学时总结】 ◆学时·II◆ IDA*算法
[学时·II] IDA*算法 ■基本策略■ 如果状态数量太多了,优先队列也难以承受:不妨再回头看DFS-- A*算法是BFS的升级,那么IDA*算法是对A*算法的再优化,同时也是对迭代加深搜索(IDF ...
- DNA sequence(映射+BFS)
Problem Description The twenty-first century is a biology-technology developing century. We know tha ...
随机推荐
- C#.net磁盘管理以及文件操作
原文发布时间为:2008-08-08 -- 来源于本人的百度文章 [由搬家工具导入] 需要引入的命名空间: using System.IO;using System.Text; private ...
- msp430项目编程34
msp430中项目---flash编程34 1.电路工作原理 2.代码(显示部分) 3.代码(功能实现) 4.项目总结
- ajax 提交数组,mybatis 数组接受
function jinxingzhong() { // 配送单编号:psd2017090103 var divHtml = ""; $.ajax({ type: "PO ...
- SGU104 二维dp
大致题意: n个东西放在(1.2.3...m)个容器中,先放的必需在后方的左边.a[i][j]表示i号物品放在j容器所得 的价值,求最大价值. 几乎是刚刚开始接触动态规划题,开始我这样想 每个东西一件 ...
- win10+Linux18.04双系统安装
给好多可爱的妹子重装了那么多次电脑,懒得码过程,因为我一般每次都要查一查...这次来个综合版吧,超简单,无脑操作. 首先说一下我的电脑Thinkpad + 500G 硬盘 (2014年买的老电脑) 首 ...
- php 处理大文件方法 SplFileObject
$csv_file = 'tmp.csv'; $start = isset($_GET['start']) ?intval($_GET['start']) : 1; // 从第几行开始读取 $num ...
- centos 複製時顯示進度的指令 pv
Pipe Viewer 的简称pv:意思是通过管道显示数据处理进度的信息.这些信息包括已经耗费的时间,完成的百分比(通过进度条显示),当前的速度,全部传输的数据,以及估计剩余的时间. yum inst ...
- SolidEdge 如何绘制零件图的剖视图
1 点击检视-剖面,然后选择剖切面 2 比如要全剖,则绘制好方框之后点返回,选择方向. 选择剖切深度,然后预览即可 一个零件可以进行多次剖切
- lua 获取当前执行目录 lfs 库
lua lfs 库 lfs.attributes(filepath [, aname]) 获取路径指定属性 lfs.chdir(path) 改变当前工作目录,成功返回true,失败返回nil加上错误信 ...
- 生成可重集的排序 (白书P184)
#include<iostream> #include<cstdio> #include<cstring> #include<algorithm> us ...
